


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 6-12-2015
Date: 21-11-2020
Date: 17-4-2021
|
DNA Topology
Double-stranded acquire DNA structures topological properties if organized into a topological domain, which is a closed region in which the two strands are linked. The known types of elementary topological domains are closed circular duplex DNA (cdDNA), in which the two strands are separately covalently continuous; and protein-sealed closed DNA loops (Fig. 1) (1). The vast majority of experimental and theoretical work on DNA topological domains has been done with purified closed circular DNA. An elementary topological domain, in which the DNA is topologically monomeric, is one in which the axis of the DNA is not itself linked to that of any other DNA. DNAs of this type are widespread in nature, including bacterial plasmids and episomes, the virion and replicative forms of both eucaryotic and procaryotic viruses, mitochondrial DNA, and many others (2). The nomenclature used to describe these molecules derives in part from protein chemistry and is imprecise when applied to closed circular DNA. Among the terms commonly used are superhelical DNA and supercoiled DNA. These are not meant to indicate different structural types, but are used as synonyms to indicate that the DNA in question is more compact than either its relaxed closed circular or nicked circular counterpart. They are used interchangeably with the more generic closed circular DNA or, alternatively, closed duplex DNA (cdDNA).

Figure 1. Depiction of the two kinds of elementary topological domains. (a) The structure of a closed duplex DNA (cdDNA), shown here as a relaxed duplex (rdDNA). Both strands in this DNA are covalently continuous, and the linking number is an integer. There are eight nodes in the projection shown here, and Lk = +4. (b) The structure of a DNA loop. The topological domain is maintained by tight binding to a dimeric protein. In the example shown, Lk≈6. The exact value of Lk depends on the precise location at which the two strands are fixed together by the protein sealant, and Lk can consequently be a fractional number. (Reproduced with permission from Ref. 1; copyright 1986, Macmillan Publishing Co.)
Protein-sealed DNA loops can occur locally in parts of DNA molecules that are not necessarily closed circular. Examples of these are the genome of Escherichia coli, which is organized into separate topological domains by bound proteins (3, 4), the lambda repressor (5, 6), several prokaryotic transcription regulation systems (7), site-specific recombination and transposition (8), and the binding of the SfiI restriction enzyme (9). It should be noted that protein-sealed DNA loops, in contrast to cdDNAs, are held together by noncovalent bonds. The integrity of this type of topological domain is therefore relatively less certain. Systematic determinations of the stability of DNA loops as a function of linking number have not yet been reported.
DNA can also be organized into higher order (nonelementary) topological domains, in which the duplex DNA axes of one or more circular DNAs are themselves linked. Knotted DNA, one of the possible outcomes of site-specific recombination, represents a class in which the duplex axis of a single DNA forms a knot. Catenated DNA, in which the axes of two or more DNA submolecules are linked, contains one or more higher order topological domains. The submolecules themselves may or may not contain individual elementary topological domains. Schematic depictions of DNA knots and catenanes for some simple cases are shown in Figure 2 (10).

Figure 2. Two kinds of higher order topological domains. (a) Two closed circular, but not supercoiled, DNAs are linked once to form a catenane. Here the higher order linking number (called the catenation number in this case) is 1, since the axes of the two DNAs are linked once. (b) The axis of a single DNA molecule is knotted into a trefoil.
The formation of an elementary topological domain has a major influence on many DNA properties and, in addition, creates some properties that are entirely new. The properties modified may be roughly divided into two classes: structure and energetics. The principal features of DNA structure affected are the duplex twist and the disposition in space of the DNA axis. Neat closed circular DNAs tend to form fairly regular, often branched superhelices in solution. A topological domain has a specific free energy, called either the topological domain free energy or the free energy of superhelix formation. This quantity generally increases with the extent of supercoiling of either chirality. The equilibria of all binding reactions that involve changes in either the DNA twist or in the writhe are shifted by the inclusion of this free-energy term. Examples are the binding of intercalative drugs, such as ethidium bromide, and of proteins, such as RNA polymerase.
The new property created by the formation of a closed circular DNA is the linking number, Lk, which is a measure of how many times the two duplex strands are interwound. It is constant for a given topological domain and can be changed only by one or more chain scissions. This makes possible the formation of DNA topoisomers , which are closed circular DNAs that differ only in Lk. Topoisomers can be reversibly interconverted with enzymes known as topoisomerases. The properties of individual topoisomers depend strongly on the magnitude of the associated linking number. Topoisomers can, under appropriate conditions, be physically separated by means of gel electrophoresis. Figure 3 (11) shows such a fractionation for a representative closed circular DNA.
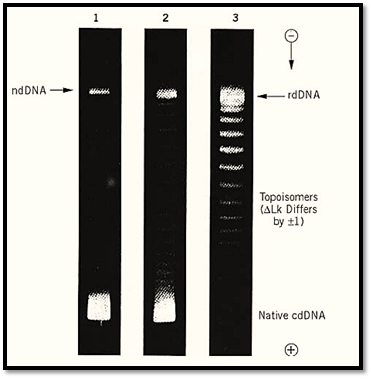
Figure 3. An example of the separation of DNA topoisomers by electrophoresis in an agarose gel. The three strips show the stepwise relaxation of closed circular SV40 DNA at 0° by incubation with a topoisomerase. Lane 1 shows the original mixture of superhelical (fastest band, labeled “Native cdDNA”) and nicked circular (slowest band, labeled “ndDNA”) species. Lanes 2 and 3 show the formation of topoisomers following 5 min and 30 min of incubation. The lowest energy product DNA is labeled “rdDNA,” and the starting superhelical DNA had DLk = –23. The individual bands contain topoisomers that differ by unity in linking number. (Because of limited gel resolution at higher DLk, not all of the higher topoisomers can be seen in this gel.)
Finally, the two DNA backbone strands whose interwinding defines a topological domain contain no chemical bonds in common (except for a possible indirect connection via a protein sealant).
Nonetheless, the two strands can be separated only by the breakage of a covalent bond. This type of connection is sometimes termed a topological bond.
References
1. M. Ptashne (1986) Nature 322, 697–701.
2. W. R. Bauer (1978) Ann. Rev. Biophys. Bioeng. 7, 287–313.
3. A. Worcel and E. Burgi (1972) J. Mol. Biol. 71, 127–147.
4. P. Sloof, A. Maagdelijn, and E. Boswinkel (1983) J. Mol. Biol. 163, 277–297.
5. J. Griffith, A. Hochschild, and M. Ptashne (1986) Nature 322, 750–752.
6. A. Hochschild and M. Ptashne (1986) Cell 44, 681–687.
7. A. Hochschild (1990) In DNA Topology and its Biological Effects (N. R. Cozzarelli and J. C. Wang, eds.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 107–138.
8. G. Chaconas, B. D. Lavoie, and M. A. Watson (1996) Curr. Biol. 6, 817–820.
9. L. M. Wentzell and S. E. Halford (1998) J. Mol. Biol. 281, 433–444.
10. A. Maxwell and M. Gellert (1986) Adv. Protein Chem. 38, 69–107.
11. W. Keller and I. Wendel (1975) Cold Spring Harb. Symp. Quant. Biol. 39 (Pt. 1), 199–208.



|
|
|
|
اكتشاف تأثير صحي مزدوج لتلوث الهواء على البالغين في منتصف العمر
|
|
|
|
|
|
|
زهور برية شائعة لتر ميم الأعصاب التالفة
|
|
|
|
|
|
جمعيّة العميد وقسم الشؤون الفكريّة تدعوان الباحثين للمشاركة في الملتقى العلمي الوطني الأوّل
|
|
|
|
الأمين العام المساعد لجامعة الدول العربية السابق: جناح جمعية العميد في معرض تونس ثمين بإصداراته
|
|
|
|
المجمع العلمي يستأنف فعاليات محفل منابر النور في واسط
|
|
|
|
برعاية العتبة العباسيّة المقدّسة فرقة العبّاس (عليه السلام) تُقيم معرضًا يوثّق انتصاراتها في قرية البشير بمحافظة كركوك
|