


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 17-5-2016
Date: 14-12-2015
Date: 29-11-2015
|
DNA Polymerase Sliding Clamps
Chromosomal replicases are multiprotein assemblies composed of a DNA polymerase and several accessory proteins and are characterized by remarkable speed and processivity. The term “processivity” refers to the fact that the polymerase is capable of polymerizing thousand of nucleotides without dissociating from the DNA. In several well-characterized systems (bacteria, archaea, eukarya, and bacteriophage-T4), the replicases have been shown to be similar in function, structure, and overall organization, and in all these systems the remarkable processivity is achieved, in party, by the DNA polymerase accessory proteins. The polymerase accessory proteins can be divided into two group: a complex of proteins known as the “clamp loader,” and a processivity factor called the “sliding clamp” (1). The sliding clamp is a ring-shaped protein (Fig. 1) that encircles DNA and anchors the polymerase to the DNA template (Fig. 2). The sliding clamp cannot assemble itself around DNA but is loaded onto DNA by the clamp loader. The arrangement of the polymerase and sliding clamp allows for rapid and processive DNA synthesis (Fig. 2). The sliding clamps have also been shown to play a role in DNA repair and gene transcription and to interact with cellular components involved in progression and control of the cell cycle.
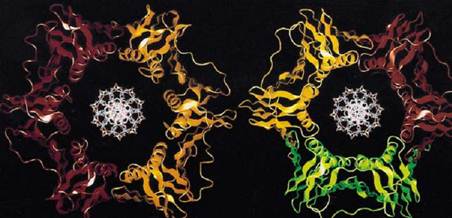
Figure 1. Computer generated images of E. coli, b subunit, and yeast PCNA.
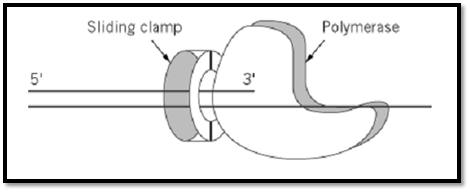
Figure 2. A model for the action of a sliding clamp as a processivity factor. The sliding clamp encircles the DNA and bin the polymerase, thus tethering it to the DNA for processive DNA synthesis.
1. Structure and Biochemical Properties of the Sliding Clamp
The sliding clamps of bacteria, archaea, eukaryotes and phage-T4 are similar in both structure and function (2-4). In Escherichia coli, the sliding clamp is the b subunit; in archaea and eukaryotes, it is the product of the proliferating cell nuclear antigen (PCNA) gene, and in phage-T4, it the product of gene-45 (gp45). The b subunit is active as a homo-dimer, whereas PCNA and gp45 are active as home-trimers (Fig. 1) (3, 5). The molecular mass of the dimer of the b subunit is 81 kDa (40.5 kDa as monomer), the PCNA trimer is 87 kDa (27 kDa as monomer), and gp45 trimer is 74 kDa (24.5 kDa as monomer) (6). Thus, the molecular mass of these oligomers are similar, and it would be shown that the b subunit, PCNA, and gp45 have similar structures.
1.1. The Three-Dimensional Structure
The processivity factors are ring-shaped proteins that encircle DNA. The overall structure of these sliding clamps are very similar; the PCNA and the b subunit rings are superimposable (Fig. 1) (5, 7-12). Each ring has similar dimensions and a central cavity large enough to accommodate a double-stranded DNA (dsDNA) molecule (Fig. 1) ((7-12); reviewed in: (3-5)). Although PCNA and gp45 are trimers and the b protein is a dimer, they each have sixfold symmetry. The symmetry derives from the two (PCNA and gp45) or three (the b subunit) globular domains, each within each monomer that have the same chain fold. Although the domains in each monomer do not have significant sequence similarity, they are superimposable in three dimensions (7). Each domain contains two a-helices that line the central cavity and are perpendicular to the DNA. The a-helices are supported by a continuous layer of b-sheet structures all around the outside that also form the intermolecular boundaries (7-12).
The sliding clamps are quite acidic proteins and would be repelled by DNA. The charge distribution on the ring, however, is asymmetric. The outer surface has strong negative electrostatic potential, which may prevent the clamp from nonspecific interactions with DNA. On the other hand, the inside of the central cavity, where the DNA is located, has a net positive electrostatic potential and, thus, may strengthen the interaction with DNA upon proper assembly of the ring around by the clamp loader.
Although the three prototypic clamps have a similar structure, they differ in their relative stability, in solution and on DNA. The b subunit forms a stable dimer in solution and is also stable on DNA while PCNA is not as stable. On the other hand, gp45 is much less stable in solution and on DNA (6).
2. The Biochemical Properties
The first indication for the toroidal shape of the sliding clamps came from the study of the b subunit of the E. coli replicase. The b subunit was shown to bind tightly to circular DNA but dissociate readily upon linearization of the plasmid by sliding off the ends (6, 13). This dissociation of b from linear DNA could be prevented if the DNA ends were blocked (6, 13). Because the affinity of b to DNA depended on the geometry of the DNA molecule, these observations suggested a toroidal shaped structure for the sliding clamp and a topological mode of binding.
Evidence for the topological binding of PCNA to DNA came from the observation that PCNA alone can support processive replication by the polymerase in the absence of clamp loader only when the DNA is linear and has a double-stranded end (14). These results demonstrated that PCNA could thread itself onto the end of the dsDNA and slide along the duplex until it reaches the 3′terminus where it interacts with the polymerase to initiate processive DNA synthesis (Fig. 2). Photocrosslinking experiments also demonstrated the sliding property of PCNA. PCNA can be crosslinked to DNA after its assembly around circular DNA. This crosslinking did not occur if the DNA was linearized, suggesting that the PCNA had dissociated from the DNA via sliding over the ends (15).
Cryoelectron microscopy was used to observe gp45, which appeared to encircle DNA; the results also indicated that it might slide along the duplex (16). The ability of gp45 to slide along DNA was confirmed using photochemical cross-linking analysis. gp45 could be cross-linked to circular, but not linear, DNA, suggesting that the protein could slide offer linear DNA (15).
3. Functions of the Processivity Factors
3.1. Processivity Factors and DNA Replication
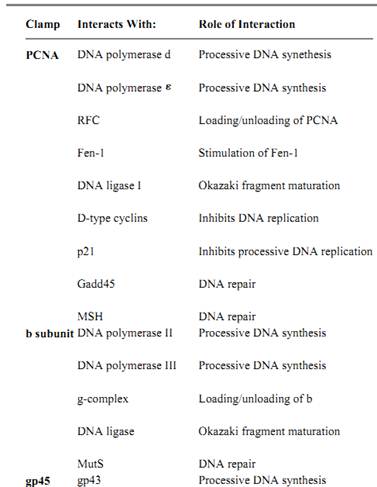
To data, the best understood function for the sliding clamps is their essential role in chromosomal DNA replication. The processivity factors endows its cognate polymerase with the ability to polymerize thousands of nucleotides without dissociating from the DNA template. The b subunit is the processivity factor of DNA polymerase (pol) III holoenzyme; PCNA is a part of the pold holoenzyme, and gp45 binds the product of gene-43 (phage-T4 polymerase).
In bacteria the processivity factor is essential for DNA replication. dnaN is the gene that encodes the E. coli b subunit (17). E. coli cells harboring a temperature-sensitive mutation in the dnaN gene stop DNA synthesis when shifted to the restrictive temperature (18). This observation, together with early biochemical analysis (19), indicated a role for the b subunit in DNA replication. Further studies demonstrated that the b subunit is responsible for the processivity of both DNA polII and polIII (Table 1). The polymerase of the E. coli polIII holoenzyme incorporates approximately 20 nucleotides/s and is processive for only 11 nucleotides before dissociating from the DNA template. Upon binding of the b subunit, polIII becomes a very rapid (750 nucleotides/s) and highly processive (>50,000 nucleotides) enzyme (20). Upon the assembly of the b subunit around the DNA template by the accessory complex, the clamp loader can be removed, leaving the clamp around the DNA. Then, the b ring can associate with the polymerase (Fig 2) to initiate rapid and processive DNA synthesis (21). Thus, at least in prokaryotes, the formation of a processive holoenzyme can be seen as a two step process. In the first step, a b clamp is assembled around the DNA primer by the clamp loader to form the “preinitiation complex.” In the second step, the polymerase assembles with the preinitiation complex to form the “initiation complex” capable of rapid and processive DNA synthesis (see Clamp Loaders of DNA Polymerases). Therefore, the mechanism by which a b ring confers processivity to polymerase is by direct contact with it, tethering the polymerase to the DNA (Fig. 2). The polymerase would then pull b along during polymerization.
Table 1. Partial List of Sliding Clamp Interacting Proteins

In eukaryotes, the early indication of a role for PCNA in DNA synthesis came from the pattern of expression during the cell cycle. PCNA is a nuclear protein with elevated expression during the S-phase (DNA synthesis) of the cell cycle (22, 23) whereas in quiescent and senescent cells there are very low levels of the protein.
Several in vivo and vitro studies determined the important role played by PCNA in DNA replication. Expression of anti-sense RNA in exponentially growing cells caused a suppression of DNA replication and cell-cycle progression (24). Deletion of the PCNA gene in both the budding yeast (25) and the fission yeast (26) demonstrated that PCNA is an essential protein that is important for DNA replication. In vitro replication assays have been employed to establish the role of PCNA as the processivity factor of Pold (Table 1), the replicase of eukaryotic cells. PCNA enables pold to replicate a template with long single-stranded stretches (27) and is required for Simian virus 40 (SV-40) in vitro replication (28), while antibodies generated against PCNA inhibit pold-dependent in vitro replication (29). PCNA also plays an important role in coordinating leading and lagging strand synthesis (30).
PCNA also stimulates the activity of pol (Table 1) (31). The precise role of pol in chromosomal DNA synthesis, however, is not yet clear; it may play a role in Okazaki fragment maturation. Pol was shown to be important for DNA repair (23, 32), and its stimulation by PCNA might be important in this process.
The role of PCNA in chromosomal DNA synthesis might not be limited to its effect on the polymerase. PCNA was shown to stimulate the activity of Fen-1 and DNA ligase I (Table 1), proteins that are important for Okazaki fragment maturation on the lagging strand (33). Thus, not only does the processivity factor play an important role in DNA synthesis but also in the final steps leading to the formation of a mature duplex DNA. PCNA may also interact with other members of the replication machinery, although these interactions have not yet been elucidated.
Early studies using temperature-sensitive mutants of gp45 (and gp44/62, its clamp loader) demonstrated its importance for DNA replication (34). Further studies using purified protein in in vitro replication assays have been used to establish the role of gp45 as the processivity factor for phage-T4 polymerase (gp43) (35).
3.2. Processivity Factors and DNA Repair
Both b and PCNA are also involved in DNA repair, at least in part, via the stimulation of their respective polymerases (32, 36). As discussed above, the b subunit forms a dimeric ring that functions as the processivity factor of polII and PolIII, both of which are involved in DNA repair processes (36, 37). It was also shown that the protein interacts with mutS, a protein needed for mismatch repair (36-38). It was also demonstrated that exposure of E. coli cells to UV-irradiation induces a shorter form of b, called b* (39). b*, comprised of the C-terminal 2/3 of normal b, was shown to behave as a trimer, presumably forming a trimeric ring, and was found to stimulate DNA synthesis by polIII. Thus, b* might serve as an alternative clamp in DNA repair processes. In eukaryotes, PCNA is the processivity factor of pol , an enzyme that has been implicated in DNA repair (23, 32). PCNA becomes localized in the cell nucleus after UV-irradiation of cells not in S-phase (40); PCNA transcription is stimulated upon UV-irradiation (41), and several mutant forms of PCNA have been shown to be defective in DNA repair processes (42). These in vivo observations are consistent with in vitro repair assays that demonstrated a role for PCNA in DNA excision-repair (43, 44 reviewed in: 32, 36).
PCNA may also be involved in DNA repair in mechanisms that do not involve the polymerase (23). Upon DNA damage, there is an elevation in the level of several genes. Two of these genes, p21 and Gadd45, have been shown to bind PCNA (Table 1) (33). The role of the interactions between PCNA and these proteins in DNA repair is not yet fully understood. It was shown, however, that p21-PCNA interaction inhibits DNA replication, thus linking cell-cycle control processes to the DNA replication machinery (23).
3.3. Transcription and Other Processes
Several well characterized examples suggest a role for viral-encoded sliding clamps in RNA transcription. It was demonstrated that gp45 serves as a transcriptional activator of phage-T4 late genes (45, 46). Two other viral-encoded proteins are also involved in this mechanism: the gene-33
product (gp33) is a transcriptional coactivator, and the gene-55 product (gp55) recognizes the late promoter. The gp45 protein, in association with gp33 and gp55, binds to the E. coli RNA polymerase and directs it to the promoters of the phage genes expressed in late stages of infection (45-47) (Table 1). The clamp loader is also involved in this process as it was demonstrated that gp45 has to be assembled around DNA in order to activate transcription (46).
At least one eukaryotic virus has been shown to utilize a sliding clamp for the regulation of gene transcription. A PCNA homologue from an insect virus has been shown to be important for the expression of several viral genes. This PCNA, however, is not essential for viral DNA replication (48).
These two examples imply that cellular-encoded sliding clamps (b and PCNA) may have a similar function and may play a role in gene transcription. Upon completion of DNA replication, some rings may be left around the duplex DNA (49). These rings perhaps then bind to RNA polymerase, directly or indirectly, and regulate gene expression.
PCNA may also play a role in chromatin remodeling. Studies conducted in starfish and Drosophila have indicated a function for PCNA in the assembly of chromatin (50, 51).
4. Concluding Remarks
The DNA polymerase sliding clamp encircles DNA and endows the polymerase with its high processivity by tethering the polymerase to the DNA. In the last several years, however, it has become apparent that the sliding clamps play diverse roles in nucleic acid metabolism (e.g., DNA repair and transcription). The proteins were also shown to be involved in cell-cycle progression via their interaction with diverse cellular proteins (23). The list of proteins that interact with the clamp is not complete. Future studies will elucidate the scope of processes in which the sliding clamps are involved.
The first proteins found to have a ring-shaped structure and encircle DNA were the DNA polymerase sliding clamps. Since that structure was demonstrated for the sliding clamps, other ring-shaped protein complexes that encircle DNA have been shown to play a role in DNA metabolism (52, 53). It will be interesting to know how general this structure is, how many other proteins have a similar structure, and how many other cellular processes utilize these proteins.
References
1. C.-C. Huang, J. E. Hearst, and B. M. Alberts (1981) J. Biol. Chem. 256, 4087–4094.
2. B. Stillman (1994) Cell 78, 725–728.
3. M. M. Hingorani and M. O''Donnell (2000) Curr. Biol. 10, R25–R29.
4. M. M. Hingorani and M. O''Donnell (2000) Curr. Organic Chem. 4, 887–913.
5. Z. Kelman and M. O''Donnell (1995) Nucleic Acids Res. 23, 3613–3620.
6. N. Yao, J. Turner, Z. Kelman, P. T. Stukenberg, F. Dean, D. Shechter, Z.-Q. Pan, J. Hurwitz, and M. O''Donnell (1996) Genes to Cell 1, 101–113.
7. X. -P. Kong, R. Onrust, M. O''Donnell, and J. Kuriyan (1992) Cell 69 425–37.
8. T. S. R. Krishna, X.-P. Kong, P. M. Burgers, and J. Kuriyan (1994) Cell 79 1233–1243.
9. J. M. Gulbis, Z. Kelman, J. Hurwitz, M. O''Donnell, and J. Kuriyan (1996) Cell 87, 297–306.
10. Y. Shamoo and T. A. Steitz (1999) Cell 99, 155–156.
11. I. Moarefi, D. Jeruzalmi, J. Turner, M. O''Donnell, and J. Kuriyan (2000) J. Mol. Biol. 296, 1215–1223.
12. S. Matsumiya, Y. Ishino, and K. Morikawa (2001) Prot. Sci. 10, 17–23.
13. P. T. Stukenberg, P. S. Studwell-Vaughan, and M. O''Donnell (1991) J. Biol. Chem. 266, 11328–11334.
14. P. M. Burgers and B. L. Yoder (1993) J. Biol. Chem. 268, 19923–19926.
15. R. L. Tinker, G. A. Kassavetis, and E. P. Geiduschek (1994) EMBO J. 13, 5330–5337.
16. E. P. Gogol, M. C. Young, W. L. Kubasek, T. C. Jarvis, and P. H. von Hippel (1992) J. Mol. Biol. 224, 395–412.
17. P. M. J. Burgers, A. Kornberg, and Y. Sakakibara (1981) Proc. Natl. Acad. Sci. USA 78, 5391–5395.
18. Y. Sakakibara and T. Mizukami (1980) Mol. Gen. Genet. 178, 541–553.
19. S. H. Wickner (1978) Ann. Rev. Biochem. 47, 1163–1191.
20. M. Mok and K. J. Marians (1987) J. Biol. Chem. 262, 16644–16654.
21. Z. Kelman and M. O''Donnell (1995) Ann. Rev. Biochem. 64, 171–200.
22. R. Bravo and J. E. Celis (1980) J. Cell. Biol. 84, 795–802.
23. Z. Kelman (1997) Oncogene 14, 629–640.
24. D. Jaskulski, J. K. DeRiel, W. E. Mercer, B. Calabbetta, and R. Baserga (1988) Science 240, 297–304.
25. G. A. Bauer and P. M. J. Burgers (1990) Nucleic Acids Res. 18, 261–265.
26. N. H. Waseem, K. Labib, P. Nurse, and D. P. Lane (1992) EMBO J. 11, 5111–5120.
27. C.-K. Tan, C. Castillo, A. G. So, and K. M. Downey (1986) J. Biol. Chem. 261, 12310–12316.
28. G. Prelich, M. Kostura, D. R. Marshak, M. B. Mathews, and B. Stillman (1987) Nature 326, 471–475.
29. C.-K. Tan, K. Sullivan, X. Li, E. M. Tan, K. M. Downey, and A. G. So (1987) Nucleic Acids Res. 15, 9299–9308.
30. G. Prelich and B. Stillman (1988) Cell 53, 117–126.
31. S.-H. Lee, Z.-Q. Pan, A. D. Kwong, P. M. J. Burgers, and J. Hurwitz (1991) J. Biol. Chem. 266, 22707–22717.
32. R. D. Wood (1996) Ann. Rev. Biochem. 65, 135–167.
33. Z. Kelman and J. Hurwitz (1998) Trends Biochem. Sci. 23, 236–238.
34. R. H. Epstein, A. Bolle, C. M. Steinberg, E. Kellenberger, E. Boy de la Tour, R. Chevalley, R. S. Edgar, M. Susman, G. H. Denhardt, and A. Lielausis (1963) Cold Spring Harbor Symp. Quant. Biol. 28, 375–394.
35. N. G. Nossal and B. M. Alberts (1983) Bacteriophage T4. (C. K. Mathews, E. M. Kutter, G.
Mosig, and P. B. Berget eds.). American Society for Microbiology, Washington DC, pp. 71–81.
36. A. Sancar (1996) Ann. Rev. Biochem. 65, 43–81.
37. Z. Livneh, O. Cohen-Fix, R. Skaltier, and T. Elizur (1993) CRC Crit. Rev. Biochem. Mol. Biol. 28, 465–513.
38. F. J. Lopez de Saro and M. O''Donnell (2001) Proc. Natl. Acad. Sci. 98, 8376–8380.
39. R. Skaliter, T. Paz-Elizur, and Z. Livneh (1996) J. Biol. Chem. 271, 2478–2481.
40. J. E. Celis and P. Madsen (1986) FEBS Lett. 209, 277–283.
41. X.-R. Zeng, Y. Jiang, S.-J. Zhang, H. Hao, and M. Y. W. T. Lee (1994) J. Biol. Chem. 269, 13748–13751.
42. R. Ayyagari, K. J. Impellizzeri, B. L. Yoder, S. L. Gary, and P. M. J. Burgers (1995) Mol. Cell. Biol. 15, 4420–4429.
43. A. F. Nichols and A. Sancar (1992) Nucleic Acid Res. 20, 2441–2446.
44. M. K. K. Shivji, M. K. Kenny, and R. D. Wood (1992) Cell 69, 367–374.
45. D. R. Herendeen, G. A. Kassavetis, J. Barry, B. M. Alberts, and E. P. Geiduschek (1989) Science 245, 952–58.
46. E. P. Geiduschek (1995) Semi. Virol. 6, 25–33.
47. E. N. Brody, G. A. Kassavetis, M. Ouhammouch, G. M. Sanders, R. L. Tinker, and E. P. Geiduschek (1995) FEMS Microb. Lett. 128, 1–8.
48. D. R. O''Reilly, A.M. Crawford, and L. K. Miller (1989) Nature 337, 606.
49. A. Yuzhakov, J. Turner, and M. O''Donnell (1996) Cell 86, 877–886.
50. A. Nomura (1994) J. Cell Sci. 107, 3291–3300.
51. D. S. Henderson, S. S. Banga, T. A. Grigliatti, and J. B. Boyd (1994) EMBO J. 13, 1450–1459.
52. M. M. Hingorani and M. O''Donnell (2000) Nat. Rev. Mol. Cell Biol. 1, 22–30.
53. M. M. Hingorani and M. O''Donnell (1998) Curr. Biol. 8, R83–R86.



|
|
|
|
هذه العلامة.. دليل على أخطر الأمراض النفسية
|
|
|
|
|
|
|
إحصائية مركز الوارث (ديرمان) للأطراف الذكية والتأهيل الطبي التابع لهيئة الصحة في العتبة الحسينية للعام (2024)
|
|
|