


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 8-12-2015
Date: 2-5-2016
Date: 24-4-2021
|
Blot Overlays
Regardless of whether a blot contains immobilized DNA, RNA or protein (see Blotting), it is ultimately reacted with a specific probe that defines the type of ligand-binding interaction being studied (see Southern Blots (DNA Blots), RNA Blots (Northern Blots), and Protein Blots (Western Blots)). Radiolabeled probes are detected by autoradiography, and the radioactive complexes are quantified by computer-digitized imaging or simply by excising out the bands and “counting” the radioactivity. Often the primary probe itself is not easily detectable, so a secondary probe is used, such as an antibody against the first probe conjugated to an enzyme, or avidin or streptavidin when the primary probe is biotinylated. When using an enzyme-conjugated probe, one must ensure that the quencher or blocking reagent used on the blot has no interfering enzymatic activity. For example, hemoglobin should not be used when horseradish peroxidase is employed as the detection system.
1. DNA Probes
Traditionally, Southern and RNA blots of DNA and RNA, respectively, are probed with single-stranded DNA or double-strand DNA that has been denatured. The probes are usually radioactive, so that any duplex formed upon hybridization of the probe to a nucleic acid on the blot is detected by autoradiography. The rationale of such experiments is to identify the position of the immobilized nucleic acid in the electrophoretogram that is complementary to the sequence of the probe, so that they hybridize by Watson-Crick base pairing. The conditions for hybridization, the stringency, are regulated according to the degree of homology and complementarity between the probe and the target (1, 2). DNA is also used to probe protein blots, a procedure that has been named “Southwestern blotting.” As expected, this type of experiment is designed to reveal specific interactions between defined DNA sequences and the DNA-binding proteins that bind them, such as transcription factors (3). DNA probing of colony or plaque blots is a routine approach in recombinant DNA cloning (4.(
2. Immunoblotting
The original application of protein blots was for identifying a particular antigen in a protein gel pattern by probing the blot with its corresponding antibody (5). Detecting the immunocomplex is often accomplished by using a secondary probe, for example, a goat antimouse IgG conjugated to an enzyme, such as horseradish peroxidase, when a murine monoclonal antibody is used for the primary probe. The sensitivity of these assays is increased by combining immunoprecipitation prior to gel electrophoresis. Thus, a crude sample of proteins is first immunoprecipitated with relevant antibodies, and the precipitated proteins are subsequently resolved by electrophoresis, blotted, and probed with a monoclonal antibody of interest. The use of antibodies to probe colony or plaque blots is very effective in screening expression libraries (6.(
3. Lectin Blotting
To identify glycoproteins, lectins can be used to probe protein blots (7). A radioactive or enzyme-conjugated lectin is employed as the probe. A particular case of interest is the fact that the enzyme horseradish peroxidase is itself a glycoprotein. Thus, for example, a blot can be probed with the mannose-specific lectin concanavalin A, washed and further incubated with horseradish peroxidase directly (8). The multivalent concanavalin A binds to blotted glycoproteins containing mannose and subsequently also binds to the horseradish peroxidase without the need for chemical conjugation. When using lectins, ensure that the quencher used does not itself contain sugar.
4. Ligand Blotting
Blotting is usually considered for detecting antigens or nucleic acid hybrids, but it is also a very powerful way to identify all sorts of protein–protein interactions, even interactions involving nonpeptide ligands. Thus when studying any receptor, one should consider probing protein blots with any corresponding ligands that are detectable (9). In such experiments, it is usually advisable not to boil the protein sample or subject it to disulfide bond reduction before electrophoresis. Furthermore, the blot can be incubated in solutions that promote renaturation of the immobilized proteins. Omission of methanol from the transfer buffers used in electroblotting is also helpful for retaining functional conformations of the blotted protein. Ligand blotting has been used successfully to identify peptides that bind to hormones, cytoskeletal components, neurotoxins, nucleotides, calmodulin, and even ions such as Ca2+ (9-11) (Fig. 1).
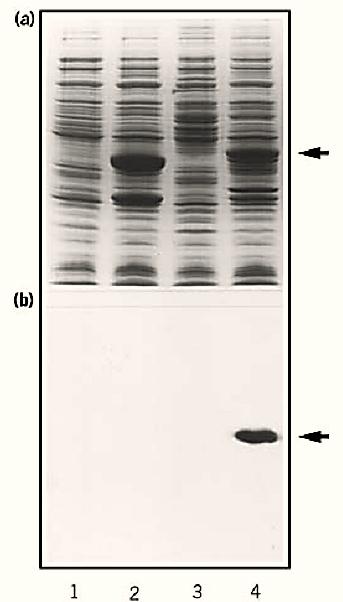
Figure 1. Neurotoxin overlays of protein blots. Escherichia coli transformed with pATH2 expression vectors containing DNA inserts corresponding to the neurotoxin binding site (lanes 3 and 4) or not (lanes 1 and 2) were induced for gene expression (lanes 2 and 4) or not (lanes 1 and 3). Then samples of all four treatments were resolved by SDS-PAGE and stained with Coomassie brilliant blue (a) or blotted onto nitrocellulose membrane filters and probed using radioactive neurotoxin. Then the washed filters were autoradiographed (b). The arrows indicate the position of the fusion protein containing the toxin binding site.
5. Cell Blotting
Blots have even been probed with intact cells, which has proven useful for identifying interactions between proteins and cells (12). Furthermore, bacteria can be used to probe a blot, and the interaction with a protein is detected by subsequently allowing the bacteria to grow on the surface of the blot, so that colonies are observed at the site of the immobilized protein (13). Viruses have also been used to probe blots to detect their corresponding receptor proteins.
have also been used to probe blots to detect their corresponding receptor proteins.
In summary, one should consider probing blots with selected ligands any time bimolecular interactions are to be analyzed. Surveying literature databases using specific conjugations, such as “ligand-blot”, “calmodulin-blot” or “cell-blot,” as key words usually produces results that provide the imaginative investigator with starting points from which to proceed.
References
1.J. Meinkoth and G. Wahl (1984) Anal. Biochem. 138, 267–284.
2.L. G. Davis, M. D. Dibner, and J. F. Battey (1986) Basic Methods in Molecular Biology, Elsevier, New York, pp. 62–65.
3.C. N. Flytzanis (1994) In Protein Blotting: A Practical Approach (B. S. Dunbar, ed.), IRL Press, Oxford, UK, pp. 163–168.
4. Ref. 2 pp. 227–229.
5. H. Towbin and J. Gordon (1984) J. Immunol. Methods 72, 313–340.
6. T. V. Huynh, R. A. Young, and R. W. Davis (1986) In DNA Cloning, A Practical Approach, Vol. 1 (D. M. Glover, ed.), IRL Press, Oxford, UK, pp. 49–78.
7.S. Bar-Nun and J. M. Gershoni (1994) In Cell Biology; a laboratory handbook, Vol 3 (J. E. Celis, ed.), Academic Press, San Diego, pp. 323–331.
8.J. C. S. Clegg (1982) Anal. Biochem. 127, 389–394.
9.J. M. Gershoni (1988) Methods of Biochemical Analysis 33, 1–58.
10. P. Hossenlopp and M. Binoux (1994) In Protein Blotting: A Practical Approach (B. S. Dunbar, ed.), IRL Press, Oxford, UK, pp. 169–188.
11. A. Vieira, R. G. Elkin, and K. Kuchler (1994) In Cell Biology: A laboratory handbook, Vol. 2 (J. E. Celis, ed.), Academic Press, San Diego, pp. 314–321.
12. E. G. Hayman, E. Engvall, E. A''Hearn, D. Barnes, M. Pierschbacher, and E. Rouslahti (1982) J. Cell Biol. 95, 20–23.
13. J. M. Gershoni (1987) Protein blotting: a tool for the analytical biochemist, Adv. Electrophoresis 1, 141–176.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|