


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 16-6-2021
Date: 20-11-2020
Date: 18-5-2016
|
Eukaryotes Use a Complex of Many Initiation Factors
KEY CONCEPTS
- Initiation factors are required for all stages of initiation, including binding of the initiator tRNA, attachment of the 40S subunit to the mRNA, joining of the 60S subunit, and movement of the ribosome along the mRNA.
- Eukaryotic initiator tRNA is a Met-tRNA that is different from the Met-tRNA used in elongation, but the methionine is not formylated as it is for the prokaryotic initiator tRNA.
- eIF2 binds the initiator Met-tRNAi and GTP, forming a ternary complex that binds to the 40S subunit before it associates with mRNA.
- A cap-binding complex binds to the 5′ end of mRNA prior to association of the mRNA with the 40S subunit.
Initiation in eukaryotes has the same general features as in prokaryotes in using a specific initiation codon and initiator tRNA. Initiation in eukaryotic cytoplasm uses AUG as the initiator codon. The initiator tRNA is a distinct type, but its methionine does not become formylated, as in prokaryotes. It is called tRNAiMet . Thus, the difference between the initiating and elongating Met-tRNAs lies solely in the tRNA portion of the complex, with Met-tRNAi used for initiation and Met-tRNAm used for elongation.
At least two features are unique to the initiator tRNAiMet in yeast: It has an unusual tertiary structure, and it is modified by phosphorylation of the 2′-ribose position on base 64 (if this modification is prevented, the initiator can be used in elongation). Thus, a distinction between initiator and elongator Met-tRNAs is maintained in eukaryotes, but its structural basis is different from that in prokaryotes.
Eukaryotic cells have more initiation factors than prokaryotic cells do: The current list includes about a dozen factors that are directly or indirectly required for initiation. The factors are named similarly to those in prokaryotes (sometimes by analogy with the bacterial factors) and are given the prefix “e” to indicate their eukaryotic origin. They act at all stages of the process, including:
-Forming an initiation complex with the 5′ end of mRNA
-Forming a complex with Met-tRNAi
-Binding the mRNA-factor complex to the Met-tRNA -factor complex
-Enabling the ribosome to scan mRNA from the 5′ end to the first AUG
-Detecting binding of initiator tRNA to AUG at the start site
-Mediating joining of the 60S subunit
Figure 22.19 summarizes the stages of initiation and shows which initiation factors are involved at each stage. eIF2, together with Met-tRNA , eIF3, eIF1, and eIF1A, binds to the 40S ribosome subunit to form the 43S preinitiation complex. eIF4A, eIF4B, eIF4E,and eIF4G bind to the 5′ end of the mRNA to form the cap-binding complex. This complex associates with 3′ end of the mRNA via eIF4G, which interacts with poly(A) binding protein (PABP). The 43S complex binds the initiation factors at the 5′ end of the mRNA and scans for the initiation codon. It can be isolated as the 48S initiation complex.
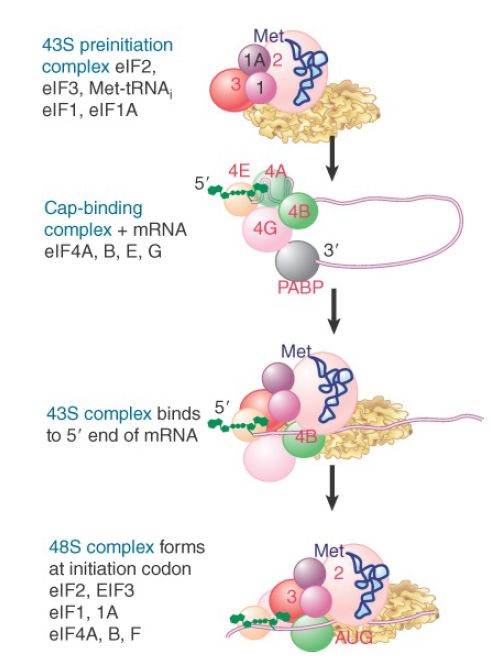
FIGURE 1.Some eukaryotic initiation factors bind to the 40S ribosome subunit to form the 43S preinitiation complex; others bind to mRNA. When the 43S complex binds to mRNA, it scans for the initiation codon and can be isolated as the 48S complex.
The subunit eIF2 is the key factor in binding Met-tRNA . Unlike prokaryotic IF2, which is a monomeric GTP-binding protein, eIF2 is a heterotrimeric GTP-binding protein consisting of α, β, and γ subunits, none of which is homologous to bacterial IF2 . eIF2 is active when bound to GTP and inactive when bound to guanine diphosphate (GDP).
Figure 2 shows that the eIF2-GTP binds to Met-tRNA . The product is sometimes called the ternary complex (after its three components, eIF2, GTP, and Met-tRNA ). Assembly of the ternary complex is regulated by the guanine nucleotide exchange factor (GEF) eIF2B, which exchanges GDP for GTP following hydrolysis of GTP by eIF2.
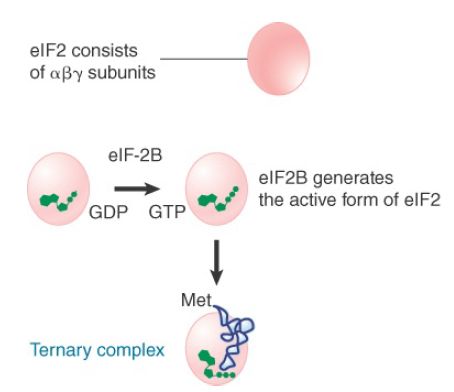
FIGURE 2 In eukaryotic initiation, eIF-2 forms a ternary complex with Met-tRNA and GTP. The ternary complex binds to free 40S subunits, which attach to the 5′ end of mRNA.
Figure 3 shows that the ternary complex places Met-tRNAi onto the 40S subunit. Along with factors eIF1, eIF1A, and eIF3, this generates the 43S preinitiation complex. The reaction is independent of the presence of mRNA. In fact, the Met-tRNAi initiator must be present in order for the 40S subunit to bind to mRNA. eIF3, which is required to maintain 40S subunits in their dissociated state, is a very large factor, with 8 to 10 subunits. eIF1 and eIF1A, which is homologous to bacterial IF1, appear to enhance eIF3’s dissociation activity.
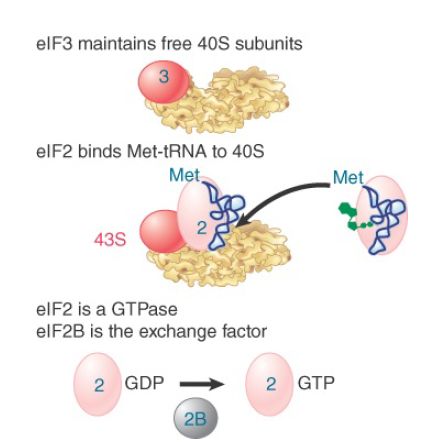
FIGURE 3. Initiation factors bind the initiator Met-tRNA to the 40S subunit to form a 43S complex. Later in the reaction, GTP is hydrolyzed and eIF2 is released in the form of eIF2-GDP. eIF2B regenerates the active form.
Figure 4 shows the group of factors that bind to the 5′ end of mRNA. The factor eIF4F is a protein complex that contains three of the initiation factors. It appears that they preassemble as a complex before binding to mRNA. The complex includes the capbinding subunit eIF4E, the helicase eIF4A, and the “scaffolding” subunit eIF4G. After eIF4E binds the cap, eIF4A unwinds any secondary structure that exists in the first 15 bases of the mRNA. Energy for the unwinding is provided by hydrolysis of ATP. Unwinding of the structure further along the mRNA is accomplished by eIF4A together with another factor, eIF4B. The main role of eIF4G is to link other components of the initiation complex.
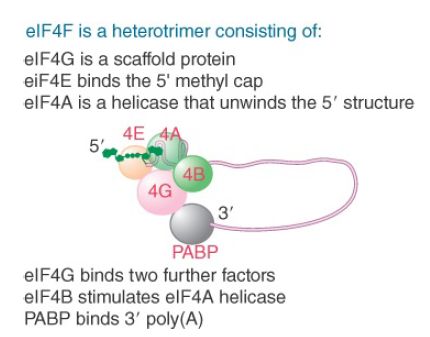
FIGURE 4. The heterotrimer eIF4F binds to the 5′ end of mRNA as well as to other factors.
The subunit eIF4E is a focus for regulation. Its activity is increased by phosphorylation, which is triggered by stimuli that increase translation and reversed by stimuli that repress translation. The subunit eIF4F has a kinase activity that phosphorylates eIF4E. The availability of eIF4E is also controlled by proteins that bind to it (called 4E-BP1, -2, and -3), to prevent it from functioning in initiation.
The presence of a poly(A) tail on the 3′ end of the mRNA stimulates the formation of the initiation complex at the 5′ end. PABP binds to the eIF4G scaffolding protein, bringing about a circular organization of the mRNA with both the 5′ and 3′ ends held in this complex. The formation of this closed loop stimulates translation; PABP is required for this effect, meaning that PABP effectively serves as an initiation factor. The PABP–eIF4G interaction on the mRNA
promotes the recruitment of the 43S complex to the mRNA, as well as the joining of the 60S subunit.
Figure 5 shows that the interactions involved in binding the mRNA to the 43S complex are not completely defined, but appear to involve eIF4G and eIF3, as well as the mRNA and 40S subunit. The subunit eIF4G binds to eIF3. This provides the means by which the 40S ribosomal subunit binds to eIF4F and thus is recruited to the complex. In effect, eIF4F functions to get eIF4G in place so that it can attract the small ribosomal subunit.
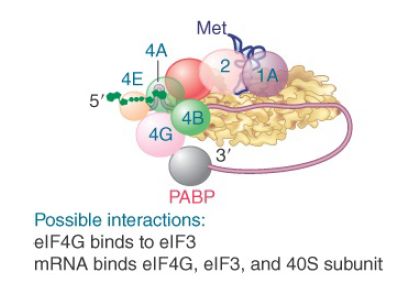
FIGURE 5. Interactions involving initiation factors are important when mRNA binds to the 43S complex.
When the small subunit has bound to the mRNA, it (usually) migrates to the first AUG codon using the Met-tRNA anticodon to find it. Scanning is assisted by the factors eIF1 and eIF1A. This process requires expenditure of energy in the form of ATP, and thus factors associated with ATP hydrolysis (eIF4A, IF4B, and eIF4F) also play a role in this step. Figure 6 shows that the small subunit stops when it reaches the initiation site, at which point the initiator tRNA base pairs with the AUG initiation codon, forming a stable 48S complex.
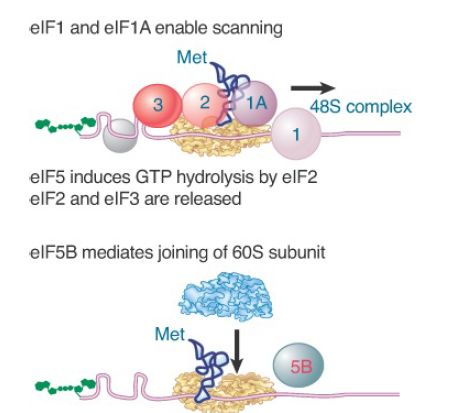
FIGURE 6. eIF1 and eIF1A help the 43S initiation complex to “scan” the mRNA until it reaches an AUG codon. eIF2 hydrolyzes its GTP to enable its release together with IF3. eIF5B mediates joining of the 60S and 40S subunits.
Joining of the 60S subunit with the initiation complex cannot occur until eIF2 and eIF3 have been released from the initiation complex. This is mediated by eIF5 and causes eIF2 to hydrolyze its GTP. The reaction occurs on the 40S subunit and requires the base pairing of the initiator tRNA with the AUG initiation codon. All of the remaining factors are likely released when the complete 80S ribosome is formed.
Finally, the initiation factor eIF5B enables the 60S subunit to join the complex, forming an intact ribosome that is ready to start elongation. eIF5B has a similar sequence to the prokaryotic initiation factor IF2, which has a similar role in hydrolyzing GTP (in addition to its role in binding the initiator tRNA). Once the factors have been released, they can associate with the initiator tRNA and ribosomal subunits in another initiation cycle. The subunit eIF2 has hydrolyzed its GTP; as a result, the active form must be regenerated. This is accomplished by the guanosine exchange factor (GEF), eIF2B, which displaces the GDP so that it can be replaced by GTP.
The subunit eIF2 is a target for regulation. Several regulatory kinases act on the α subunit of eIF2. Phosphorylation prevents eIF2B from regenerating the active form, which limits the action of eIF2B to one cycle of initiation and thereby inhibits translation.



|
|
|
|
دراسة: إجراء واحد لتقليل المخاطر الجينية للوفاة المبكرة
|
|
|
|
|
|
|
"الملح والماء" يمهدان الطريق لأجهزة كمبيوتر تحاكي الدماغ البشري
|
|
|
|
|
|
بالصور: ويستمر الانجاز.. كوادر العتبة الحسينية تواصل اعمالها في مشروع سرداب القبلة الكبير
|
|
|
|
بحضور ممثل المرجعية العليا.. قسم تطوير الموارد البشرية يستعرض مسودة برنامجه التدريبي الأضخم في العتبة الحسينية
|
|
|
|
على مساحة (150) دونما ويضم مسجدا ومركزا صحيا ومدارسا لكلا الجنسين.. العتبة الحسينية تكشف عن نسب الإنجاز بمشروع مجمع إسكان الفقراء في كربلاء
|
|
|
|
للمشاركة الفاعلة في مهرجان كوثر العصمة الثاني وربيع الشهادة الـ(16).. العتبة الحسينية تمنح نظيرتها الكاظمية (درع المهرجان)
|