


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Sigma Factor Controls Binding to DNA by Recognizing Specific Sequences in Promoters |
|
|
|
Read More
Date: 28-4-2016
Date: 30-11-2015
Date: 8-3-2021
|
Sigma Factor Controls Binding to DNA by Recognizing Specific Sequences in Promoters
KEY CONCEPTS
- A promoter is defined by the presence of short consensus sequences at specific locations.
- The promoter consensus sequences usually consist of a purine at the start point, a hexamer with a sequence close to TATAAT centered at about −10, and another hexamer with a sequence similar to TTGACA centered at about −35.
- Individual promoters usually differ from the consensus at one or more positions.
- Promoter efficiency can be affected by additional elements as well.
As a sequence of DNA whose function is to be recognized by proteins, a promoter differs from sequences whose role is to be transcribed. The information for promoter function is provided directly by the DNA sequence: Its structure is the signal. This is a classic example of a cis-acting site, as defined in the chapter titled Genes Are DNA and Encode RNAs and Polypeptides. By contrast, expressed regions gain their meaning only after the information is transferred into the form of some other nucleic acid or protein.
One way to design a promoter would be for a particular sequence of DNA to be recognized by RNA polymerase. Every promoter would consist of, or at least include, this sequence. In the bacterial genome, the minimum length that could provide an adequate signal is 12 bp. (Any shorter sequence is likely to occur—just by chance —a sufficient number of additional times to provide false signals. The minimum length required for unique recognition increases with the size of genome, a problem in eukaryotic genomes.) The 12-bp sequence need not be contiguous. If a specific number of base pairs separates two constant shorter sequences, their combined length could be less than 12 bp, because the distance of separation itself provides a part of the signal (even if the intermediate sequence is itself irrelevant). In fact, RNA polymerase recognizes promoter DNA sequences in large part from “direct readout” of specific bases in the DNA by specific amino acids in the holoenzyme. The dramatic differences in the strengths of different bacterial promoters derives in large part from variation in how well the different promoter sequences are able to be read out by the amino acid sequences present in the σ and α subunits.
Attempts to identify the features in DNA that are necessary for RNA polymerase binding started by comparing the sequences of different promoters. Any essential nucleotide sequence should be present in all the promoters. Such a sequence is said to be conserved. A conserved sequence need not necessarily be conserved at every single position, though; some variation is
permitted. How do we analyze a sequence of DNA to determine whether it is sufficiently conserved to constitute a recognizable signal?
Putative DNA recognition sites can be defined in terms of an idealized sequence that represents the base most often present at each position. A consensus sequence is defined by aligning all known examples to maximize their homology. For a sequence to be accepted as a consensus, each particular base must be reasonably predominant at its position, and most of the actual examples must be related to the consensus by only one or two substitutions.
A striking feature in the sequence of promoters in E. coli is the lack of extensive conservation of sequence over the entire 75 bp associated with RNA polymerase. Some short stretches within the promoter are conserved, however, and they are critical for its function. Conservation of only very short consensus sequences is a typical feature of regulatory sites (such as promoters) in both prokaryotic and eukaryotic genomes.
Several elements in bacterial promoters contribute to their recognition by RNA polymerase holoenzyme. Two 6-bp elements, referred to as the −10 element and −35 element (as well as the length of the “spacer” sequence between them), are usually the most important of these recognition sequences. The promoter sequence at and directly adjacent to the transcription start point, the sequences on either side of the −10 element (referred to as the extended −10 element on the upstream side and the discriminator on the downstream side), and the 10 to 20 bp directly upstream of the −35 element (referred to as the UP element), however, also interact sequence specifically with RNA polymerase and contribute to promoter efficiency:
- A 6-bp region is recognizable centered approximately 10 bp upstream of the start point in most promoters (the actual distance from the start site varies slightly from promoter to promoter). This hexameric sequence is usually called the −10 element, the Pribnow box, or sometimes the TATA box (though the latter name is preferentially applied to a similar consensus sequence in eukaryotic promoters). Its consensus, TATAAT, can be summarized in the form:
T80 A95 T45 A60 A50 T96
where the subscript denotes the percent occurrence of the most frequently found base, which varies from 45% to 96%. (A position at which there is no discernible preference for any base would be indicated by N.) The frequency of occurrence corresponds to the importance of these base pairs in binding RNA polymerase. Thus, the initial highly conserved TA and the final, almost completely conserved T in the −10 sequence are crucial for promoter recognition. It is now known that the −10 element makes sequence-specific contacts to sigma factor regions 2.3 and 2.4 (see the discussion that follows). This region of the promoter is double stranded in the closed complex and single stranded in the open complex, though, so interactions between the −10 element and RNA polymerase are complex and change at different stages in the process of transcription initiation.
- The conserved hexamer, TTGACA, centered at approximately 35 bp upstream of the start point is called the −35 element. In more detailed form, it can be written:
T82 T84 G78 A65 C54 A45
Bases in this element interact directly with region 4.2 of the sigma factor (see the discussion that follows) similarly in both the closed and open complexes.
- The distance separating the −35 and −10 sites is between 16 bp and 18 bp in about 90% of promoters; in the exceptions, it is as little as 15 bp or as great as 20 bp. Although the actual sequence in most of the intervening region is relatively unimportant, the distance is critical, because, given the helical nature of the DNA, it determines not only the appropriate separation of the two interacting regions in RNA polymerase but also the geometrical orientation of the two sites with respect to one another.
- The start point is usually (more than 90% of the time) a purine, usually adenine. It is common for the start point to be the central base in the sequence CAT, but the conservation of this triplet is not great enough to regard it as an obligatory signal.
- Certain base pairs in the region between the start point and the −10 element are contacted by region 1.2 of the sigma factor . For example, a sequencespecific interaction between a guanine residue on the nontemplate strand two positions downstream of the −10 element is especially important in determining the stability of the open complex. Thus, differences in promoter sequence at positions that are not highly conserved can contribute to the variation in the strengths of different promoters.
- Bases in the extended −10 element are contacted by region 3.0 of the sigma factor (see the discussion that follows). The sequence TGN at the upstream end of the −10 element results in interactions that are especially essential for transcription initiation when the promoter lacks a −35 element sequence that closely matches the consensus. This illustrates the modularity of promoter sequences: A weak match to the consensus in one module can be compensated for by a strong match to the consensus in another.
- The approximately 20-bp region upstream of the −35 element may interact with the CTDs of the two α subunits. Effects of these interactions on promoter activity can be quite substantial, increasing transcription well over an order of magnitude for highly expressed promoters like those in rRNA genes. When these sequences closely match the consensus, this region is referred to as the UP element.
The structure of a promoter, showing the permitted range of variation from this optimum, is illustrated in FIGURE 1.
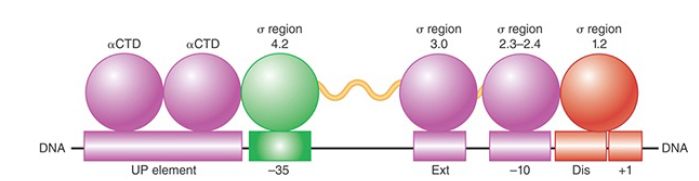
FIGURE 1. DNA elements and RNA polymerase modules that contribute to promoter recognition by sigma factor.
Data from S. P. Haugen, W. Ross, and R. L. Gourse, Nat. Rev. Microbiol. 6 (2008): 507–519.



|
|
|
|
كيف تعزز نمو الشعر الصحي؟
|
|
|
|
|
|
|
10 فحوصات مهمة يجب القيام بها لسيارتك قبل الصيف
|
|
|
|
|
|
جامعة الزهراء (عليها السلام) تكرم قسم الشؤون الفكرية بمناسبة اليوم العالمي للكتاب
|
|
|
|
قسم شؤون المعارف يقيم ندوة علمية حول جهود علماء البصرة في نشر الحديث
|
|
|
|
قسم الشؤون الفكرية يختتم برنامجاً ثقافياً لوفدٍ من جامعة البصرة
|
|
|
|
جامعة الكفيل تعقد ورشة عمل عن إجراءات عمل اللجان الامتحانيّة
|