


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 6-11-2016
Date: 29-10-2016
Date: 19-10-2016
|
Protein Synthesis
In the process of protein synthesis, ribosomes bind to mRNA and "read" its codons. Guided by the information in the nucleotide sequence of the mRNA, the ribosomes catalyze the polymerization of amino acids in the order specified by the gene from which the mRNA was transcribed.
RIBOSOMES
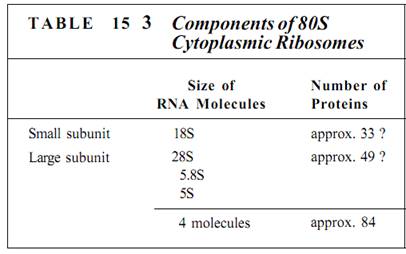
Ribosomes are small, cytoplasmic particles that "read" the genetic message in mRNA and construct proteins guided by that information. Each is composed of two subunits, one lager than the other, and each is made up of both proteins and ribosomal RNA, rRNA (Fig. 1; Table 15.3). The small subunit contains one molecule of rRNA, the large subunit one molecule each of three types of rRNA. The number of proteins present in eukaryotic ribosomes is known to be greater than 80, but the exact number is still uncertain. Ribosomes found in the cytoplasm of eukaryotes are designated 80S, meaning that they are relatively large and dense; ribosomes of plastids, mitochondria, and prokaryotes are smaller, lighter 70S ribosomes. (S is a Svedberg unit, used to measure the rate at which a particle sediments in a centrifuge.)
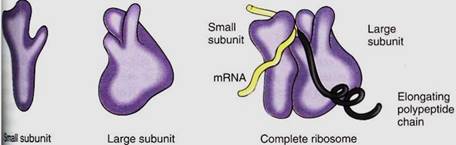
FIGURE 1:Ribosomes have two subunits, one large and one small. When they fit together there is a groove for mRNA to pass through, a channel through which the growing protein emerges, and a channel into which amino acid carriers enter. This diagram is based on ribosomes of the bacterium Escherichia coli; eukaryotic cytoplasmic ribosomes are similar, but we are not as certain about details of their shape.
The genes for the three largest rRNAs are unusual because they occur tightly grouped together and act as a single gene with just one promoter (Fig. 2). Transcription of this cluster, which is located in the nucleolus, produces a long RNA molecule that is then cut into three pieces . Short regions are digested off the ends of these pieces, resulting in three rRNA molecules. The gene for the smallest rRNA molecule (5S RNA) is not located in the nucleolus but out in the chromatin along with protein-coding genes.
Once transcribed, rRNA molecules combine with ribosome proteins in the nucleolus, forming one large particle that then is cleaved into the large and small subunits of a ribosome. These subunits are then transported out of the nucleolus through the nucleus to the cytoplasm.
All cells need large numbers of ribosomes. Most genes are present in a diploid nucleus as only two copies. But just two copies would be inadequate for producing the hundreds of thousands of rRNAs needed. Instead, rRNA genes are highly amplified; that is, many copies of each are present. In flax plants, each nucleus may have up to 120,000 copies of the gene for the small rRNA and 2700 copies of the gene cluster for the three large rRNA molecules.
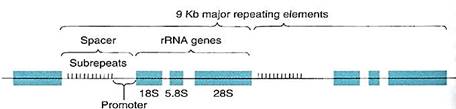
FIGURE 2:The genes for the 18S,. 5.8S, and 28S ribosomal RNAs occur together as a cluster, separated by short regions of spacer DNA. All three genes are transcribed as just one molecule of RNA, spacers included; spacer RNA is cut out, resulting in three individual rRNA molecules. Upstream from the 18S gene is the promoter and then a region made up of 10 to 15 copies of a short sequence about 135 base pairs long, called sub-repeats; they are not transcribed. On either side of the rRNA gene cluster are more rRNA clusters, as many as 2700, all occurring end to end as a gene "family." Kb is kilo-bases; 1 Kb = 1000 bases.
t RNA
During protein synthesis, amino acids are carried to ribosomes by ribonucleic acids called transfer RNA, tRNA. tRNAs are necessary because a codon cannot interact directly with an amino acid; the genetic code can be read only by a ribonucleic acid that has a three-nucleotide sequence, called an anticodon, that is complementary to and hydrogen bonds to the codon. For example, UUU and UUC are both codons for phenylalanine; the tRNAs that carry phenylalanine have the complementary anticodons of either AAA or GAA.
There are as many types of tRNA as there are codons that specify amino acids; stop codons do not have tRNAs. All tRNAs have the same parts, an anticodon and an amino acid attachment site at its 3' end consisting of the sequence CCA (Fig. 3). A special class of enzymes recognizes each tRNA and attaches the correct amino acid to it. This step, called amino acid activation, must be precise. If the wrong amino acid is placed onto a tRNA, it becomes incorporated into the protein as if it were the correct amino acid, causing the protein to have an erroneous structure.
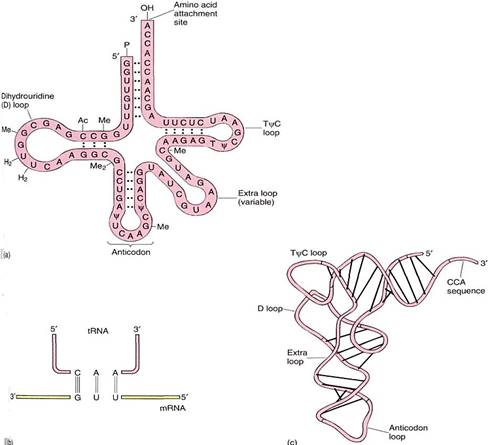
FIGURE 3: (a) and (b) All tRNAs have this general shape, with at least three loops caused by self- complementary base pairing. The small fourth loop is present only in some. The anticodon is at the middle loop and the amino acid is attached to the CCA at the 3' end. This is one of the six that carry leucine; it has the anticodon CAA, which recognizes the codon 3'-GUU-5', normally written UUG. The unusual symbols (Ac, Me, y) indicate bases that are chemically modified into unusual forms. (c) tRNA folds into an L shape rather than lying flat.
Transfer RNAs contain bases not found in other nucleotides. tRNAs are transcribed from tRNA genes, and like all ribonucleic acids, they contain the common bases A, U, G, and C. But after these are polymerized into tRNA, enzymes modify some bases to unusual forms (Fig. 3a).
Each tRNA carries an amino acid only briefly; after it is activated, it rapidly encounters a ribosome that is "reading" the codon complementary to its anticodon. It gives up its amino acid and shuttles back to the cytosol and is reactivated. Even though each tRNA can cycle like this many times per second, millions of tRNAs are needed in every cell, and hundreds or even thousands of copies of each tRNA gene may be present in each nucleus.
mRNA TRANSLATION
Initiation of Translation. The synthesis of a protein molecule by ribosomes under the guidance of mRNA is called translation. Protein synthesis begins with a complex initiation process involving the start codon AUG. This codes for the amino acid methionine, but two types of tRNA actually carry methionine, and they have different properties. One, called initiator tRNA, binds to the ribosome small subunit even though no mRNA is present (Fig. 4). Several initiation factors also bind to the small subunit. In this condition, the complex is competent to bind to mRNA, after which the large subunit of the ribosome binds to the complex and the initiation factors are released. Just how mRNA is recognized is not known, but the initiator tRNA is important for finding the AUG start codon and positioning the small subunit.
It is critically important that the ribosome be properly aligned on mRNA, because the sequence of nucleotides can be read in any set of three. The sequence CUUGCACAG can be read as CUU GCA CAG and would code for leucine alanine glutamine (Fig. 5a). But if the ribosome binds incorrectly, shifted downstream by one nucleotide, mRNA is read as C UUG CAC AG _, the codons for phenylalanine histidine and either serine or arginine, depending on the next nucleotide. Reading nucleotides in the wrong sets of three is a frameshift error; because virtually all codons are misread, frameshift errors typically result in completely useless proteins.
Elongation of the Protein Chain. Messenger mRNA lies in a channel between the two ribosome subunits. Extending outward from the mRNA channel are two grooves, each wide enough for a tRNA to fit into it such that the tRNA anticodon can touch an mRNA codon. When both channels contain activated tRNAs, adjacent codons are being read (Fig. 15.13).
At the time of initial binding, the small subunit already contains initiator tRNA in the P channel (P for protein). The adjacent A channel (A for amino acid) is empty, but numerous molecules enter it at random. Some are activated tRNAs, but if their anticodons do not complement the exposed codon at the bottom of the A channel, they diffuse out. If a tRNA with the proper, complementary anticodon enters, hydrogen-bonds form between the codon and the anticodon. This holds the tRNA in place long enough for an even more stable binding to occur. Enzymes located in the large ribosomal subunit break the bond between the methionine and its tRNA in the P channel, simultaneously attaching the methionine to the amino group of the amino acid on the tRNA in the A channel. This reaction needs no outside source of power: The broken bond is a high-energy bond, whereas the newly formed peptide bond is only a low-energy bond.
The empty tRNA, freed of its methionine, is released from the P channel, and the ribosome pulls itself along the mRNA for a distance of three nucleotides. This movement is powered by GTP and seems to be performed by proteins on the large subunit. As the ribosome slides along the mRNA, the A channel slides to the next codon and the tRNA with the two amino acids attached becomes surrounded by the P channel, not the A channel.
When the proper activated tRNA diffuses into the A channel and hydrogen bonding between codon and anticodon occurs, the large subunit enzymes again release the short protein chain (now two amino acids long) from the tRNA in the P channel and attach it to the amino acid on the tRNA in the A channel, creating a protein three amino acids long. This process repeats until the ribosome reaches a stop codon.
Termination of Translation. When a stop codon is pulled into the A channel, normal elongation cannot occur; no tRNA is present with an anticodon complementary to a stop codon. Instead, a release factor enters the channel and stimulates the large subunit enzymes to initiate the normal reactions. The high-energy bond holding the protein to the P-channel tRNA is broken, and a new bond is formed with water, releasing the protein from both the tRNA and the ribosome. The tRNA is released, and the small subunit disassociates from the large subunit and releases the mRNA. All components diffuse away, but as soon as the small subunit encounters an initiator tRNA and other initiating factors, it binds to another mRNA and the process begins again.
In summary, protein synthesis involves the following steps: RNA polymerase transcribes hnRNA, being guided by the sequence of DNA nucleotides in a gene. The hnRNA is processed into mRNA as introns are cut out and exons are spliced together. After moving from nucleus to cytoplasm, mRNA binds with ribosomes, and tRNAs carry amino acids to the ribosome-mRNA complex. The ribosome translates the mRNA codon by codon, and tRNAs fit into the ribosome only when their anticodon is complementary to the codon in the A channel. Once the protein is completed, it is released and the ribosome subunits detach from the mRNA. All components diffuse away from each other, but the ribosome subunits can translate the next mRNA they happen to meet. The mRNA can be translated again by other ribosomes.

FIGURE 4:Protein synthesis; the steps are explained in the text.

FIGURE 5:Because there are no spacers between codons, the ribonucleotides can be read in three possible ways. Each results in completely different proteins, only one of which has the proper primary structure.



|
|
|
|
5 علامات تحذيرية قد تدل على "مشكل خطير" في الكبد
|
|
|
|
|
|
|
لحماية التراث الوطني.. العتبة العباسية تعلن عن ترميم أكثر من 200 وثيقة خلال عام 2024
|
|
|