


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 23-5-2016
Date: 23-3-2021
Date: 27-2-2021
|
Eukaryotic Genes Involved in Homologous Recombination
KEY CONCEPTS
- The MRX complex, Exo1, and Sgs1/Dna2 in yeast and the MRN complex and BLM in mammalian cells resect double-strand breaks.
- The Rad51 recombinase binds to single-stranded DNA with the aid of mediator proteins, which overcome the inhibitory effects of RPA.
- Strand invasion is dependent on Rad54 and Rdh54 in yeast and Rad54 and Rad54B in mammalian cells.
- Yeast Sgs1 and Mus81/Mms4 and human BLM and MUS81/EME1 are implicated in resolution of Holliday junctions.
Previously, we briefly mentioned some of the proteins involved in homologous recombination in eukaryotes. In this section, they are discussed in more detail, focusing on the DSBR and SDSA models. Additionally, the steps in the single-strand annealing and break-induced replication mechanisms that overlap with those of DSBR and SDSA proceed by the same enzymatic processes.
Many of the eukaryotic homologous recombination genes are called RAD genes because they were first isolated in screens for mutants with increased sensitivity to X-ray irradiation. X-rays make DSBs in DNA; thus it is not surprising that rad mutants sensitive to X-rays also are defective in mitotic and meiotic recombination. The DSBR model indicates at which step the proteins described in the following paragraphs act.
1. End Processing/Presynapsis
In mitotic cells, DSBs are produced by exogenous sources such as irradiation or chemical treatment and from endogenous sources such as topoisomerases and nicks on the template strand. During replication nicks are converted to DSBs. The ends of these breaks are processed by exonucleolytic degradation to have single-strand tails with 3′–OH ends. In meiosis, DSBs are induced by Spo11-dependent cleavage. The first step in end processing entails binding of the broken end by the MRN or MRX complex, in association with the endonuclease Sae2 (CtIP in mammalian cells). Mre11 works as part of a complex with two other factors, called Rad50 and Xrs2 in yeast and Rad50 and Nbs1 in humans. Xrs2 and Nbs1 have no similarity to each other. Rad50 is thought to help hold DSB ends together via dimers connected at the tips by a hook structure that becomes active in the presence of zinc ion, as shown in FIGURE 1. Rad50 and Mre11 are related to the bacterial proteins SbcC and SbcD, which have double-stranded DNA exonuclease and single-stranded endonuclease activities. Xrs2 and Nbs1 have DNA-binding activity. Nbs1 is so named because a mutant allele was first discovered in individuals with Nijmegen breakage syndrome, a rare DNA damage syndrome that is associated with defective DNA damage checkpoint signaling and lymphoid tumors. Rare mutations that produce MRE11 with low activity have been found in humans who have ataxia-telangiectasialike disorder (ATLD). Patients with this syndrome have not been reported to be cancer prone, but they have developmental problems and show defects in DNA damage checkpoint signaling. Mutations in MRE11, RAD50, or XRS2 render cells sensitive to ionizing radiation and diploids have a poor meiotic outcome. Null mutations of MRE11, RAD50, or NBS1 in mice are lethal.
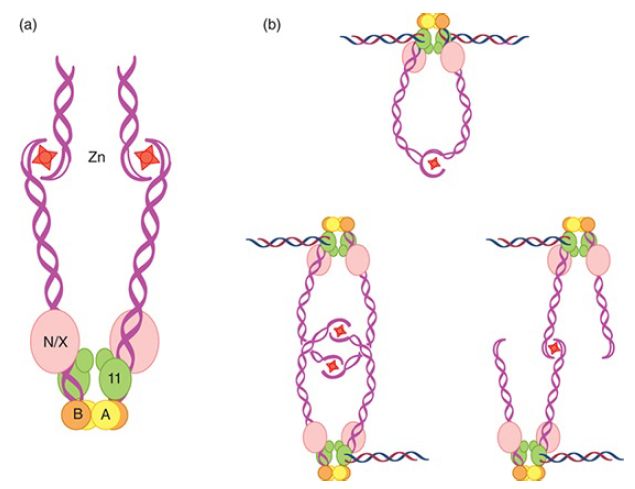
FIGURE 1 Structure of Rad50 and model for the MRX/N complex binding to double-strand breaks. Rad50 has a coiled coil domain similar to SMC (structural maintenance of chromosomes) proteins. The globular end contains two ATP-binding and hydrolysis regions (a and b) and forms a complex with Mre11 and Nbs1 (N) or Xrs2 (X). The other end of the coil binds a zinc cation and forms a dimer with another MRX/N molecule. The globular end binds to chromatin. The complex binds to double-strand breaks and can bring them together in a reaction involving two ends and one MRN/X complex (top right figure) or through an interaction between two MRX/N dimers (bottom right figure).
Data from M. Lichten, Nat. Struct. Mol. Biol. 12 (2005): 392–393.
After MRN/MRX and CtIP/Sae2 have prepared the DSB ends and removed any attached proteins or adduct that would inhibit end resection, the ends are resected by nucleases that act in concert with DNA helicases that unwind the duplex to expose single-strand DNA ends. Recent studies have identified the Exo1 and Dna2 exonucleases and the Sgs1 (in yeast) and BLM (in mammalian cells) helicases as critical factors for end processing.
After the DSBs have been processed to have 3′–OH single-strand tails, the single-strand DNA is bound first by the single-strand DNAbinding protein RPA to remove any secondary structure. Next, with the aid of mediator proteins that help Rad51 displace RPA and bind the single-strand DNA, Rad51 forms a nucleofilament. Rad51 is related to RecA with 30% identity and forms a right-handed helical nucleofilament in an ATP-dependent process, with six Rad51
molecules and 18 nucleotides of single-strand DNA per helical turn. This binding stretches the DNA by approximately 1.5-fold, compared to B-form DNA. Rad51 is required for all homologous recombination processes except single-strand annealing. RAD51 is not an essential gene in yeast, but null mutants are reduced in mitotic recombination and are sensitive to ionizing radiation. DSBs form but become degraded. In mice, RAD51 is essential, and mice
that are homozygous for mutant rad51 do not survive past early stages of embryogenesis. This is thought to reflect the fact that, in vertebrates, at least one DSB occurs spontaneously during every replication cycle as a result of unrepaired template strand nicks.
In vitro, the mediators help in the removal of RPA and in the assembly of Rad51 on the single-stranded DNA and promote in vitro strand-exchange reactions. In yeast, the mediators are Rad52 and Rad55/Rad57. Rad55 and Rad57, which form a stable heterodimer, have some homology to Rad51, but have no strandexchange activity in vitro.
In human cells, the mediators are also related to RAD51, with 20% to 30% sequence identity, and are called RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, or the “RAD51 paralogs.” (Recall that paralogs are genes that have arisen by duplication within an organism and therefore are related by sequence but have evolved to have different functions.) The human mediator proteins form three complexes: one composed of RAD51B and RAD51C, a second composed of RAD51D and XRCC2, and a third composed of RAD51C and XRCC3. The paralogous genes have been deletedin chicken cell li nes and knocked down in mammalian cells.
Although the cell lines are viable, they are subject to numerous chromosome breaks and rearrangements and have reduced viability compared to normal cell lines. Mice in which the paralogous genes have been deleted are not viable and undergo early embryonic death.
The human BRCA2 protein, which is mutated in familial breast and ovarian cancers and in the DNA damage syndrome Fanconi anemia, has mediator activity in vitro. Given that BRCA2 interacts physically with RAD51 and can bind to single-stranded DNA, this is not an unexpected activity for BRCA2. Indeed, genetic studies in mouse cells have shown that BRCA2 is required for homologous recombination. The related Brh2 protein of the pathogenic fungus Ustilago maydis binds in a complex to Rad51 and recruits it to single-strand DNA coated with RPA to initiate Rad51 nucleofilament formation.
Yeast mutants deleted for RAD55 or RAD57 show temperaturedependent ionizing radiation sensitivity and are reduced in homologous recombination. Neither mutant undergoes successful
meiosis.
Rad52 is not essential for recombination in vivo in mammalian cells and does not appear to have a mediator role in these cells. It is, however, the most critical homologous recombination protein in yeast, as rad52 null mutants are extremely sensitive to ionizing radiation and are defective in all types of homologous recombination assayed. RAD52-deficient cells never complete meiosis.
2. Synapsis
Once the Rad51 filament has formed on single-strand DNA in the DBSR and SDSA processes, a search for homology with another DNA molecule begins and, once found, strand invasion to form a Dloop occurs. Strand invasion requires the Rad54 protein and the related Rdh54/Tid1 protein in yeast, and RAD54B in mammalian cells. Rad54 and Rdh54 are members of the SWI/SNF chromatin remodeling superfamily . They possess a double-strand DNAdependent
ATPase activity, can promote chromatin remodeling, and can translocate on double-stranded DNA, inducing superhelical stress in double-stranded DNA. Although Rad54, Rdh54, and RAD54B are not DNA helicases, the translocase activity causes local opening of double strands, which may serve to stimulate Dloop formation. In yeast, RAD54 is required for efficient mitotic recombination and for repair of DSBs, because RAD54-deficient cells are sensitive to ionizing radiation and other DNA-damaging compounds. RDH54-deficient cells have a modest defect in recombination and are slightly DNA-damage sensitive. This sensitivity is enhanced when both RAD54 and RDH54 are deleted.
In meiotic cells, rad54 mutants can complete meiosis but have reduced spore viability. The rdh54 mutants are more deficient in meiosis and have a stronger effect on spore viability. The double mutant does not complete meiosis. In chicken cells and mouse cells, RAD54 and RAD54B deletion mutants are viable, in contrast to other homologous recombination gene-deletion mutants. The cells show increased sensitivity to ionizing radiation and other clastogens (agents that cause chromosomal breaks) and have reduced rates of recombination.
3. DNA Heteroduplex Extension and Branch Migration
The proteins involved in this step are not as well defined as those required in the early steps of homologous recombination, yet the homologous DSBR and SDSA recombination pathways both hav D-loop extension as an important part of the process. D-loop formation results in Rad51 filament being formed on doublestranded DNA. Rad54 protein has the ability to remove Rad51 from double-stranded DNA. This step might be important for DNA polymerase extension from the 3′ terminus. DNA polymerase delta (δ) is thought to be the polymerase for repair synthesis in DSBmediated recombination; however, some recent studies have also implicated DNA polymerase h/Rad30 as being able to extend from the strand invasion intermediate terminus.
4. Resolution
The search for eukaryotic resolvase proteins has been a long process. Mutants of the DNA helicases Sgs1 of yeast and BLM in humans result in higher crossover rates. These helicases have thus been proposed to normally prevent crossover formation by promoting noncrossover Holliday junction resolution. This is proposed to occur by branch migration of the double Holliday junctions to convergence, through the DNA helicase action, as shown in FIGURE 2. The end structure is suggested to be a hemicatenane, where DNA strands are looped around each other. This structure is then resolved by the action of an associated DNA topoisomerase: Top3 in the case of Sgs1 and hTOPOIIIα in the case of BLM. In vitro, BLM and hTOPOIIIα can dissolve double Holliday junctions into a noncrossover molecule.
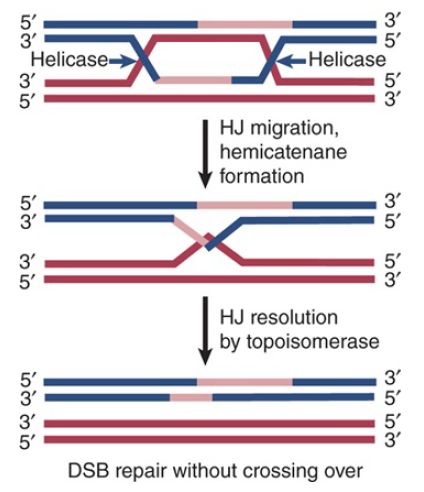
FIGURE 2. Double Holliday junction dissolution by the action of a DNA helicase and topoisomerase. The two Holliday junctions are pushed toward each other by branch migration using the DNA helicase activity. The resulting structure is a hemicatenane where single strands from two different DNA helices are wound around each other. This is cut by a DNA topoisomerase, unwinding and releasing the two DNA molecules and forming noncrossover products.
While the helicase–topoisomerase complex can resolve Holliday junctions as noncrossover in mitotic cells, the meiotic Holliday junction resolvase that can result in crossovers has not been fully identified. Additional endonuclease activities contained in the Mus81–Mms4 and Slx1–Slx4 complexes in yeast and the MUS81–EME1 and SLX1–SLX4 complexes in mammalian cells can cleave nicked Holliday junction–like structures and branched DNA structures. The relationship of this activity to meiotic crossover formation, however, is not fully defined. Recently, eukaryotic resolvase homologs were identified in humans and S. cerevisiae. The proteins GEN1 in humans and Yen1 in yeast are capable of resolving Holliday structures in vitro. These proteins are not normally essential for resolving recombination intermediates in vivo, but become essential in the absence of Mus81–Mms4.



|
|
|
|
مخاطر عدم علاج ارتفاع ضغط الدم
|
|
|
|
|
|
|
اختراق جديد في علاج سرطان البروستات العدواني
|
|
|
|
|
|
|
مدرسة دار العلم.. صرح علميّ متميز في كربلاء لنشر علوم أهل البيت (عليهم السلام)
|
|
|