


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 19-12-2015
Date: 10-5-2016
Date: 15-5-2016
|
Cyclic Amp Receptor Protein (CRP)/Catabolite Gene Activator Protein (CAP(
The cyclic AMP receptor protein (CRP), or catabolite gene activator protein (CAP), of Escherichia coli is one of the best characterized transcription factors. It was discovered more than 25 years ago as a protein that binds cyclic AMP (cAMP) and stimulates gene expression of the lac operon (1, 2). Since then, CRP has attracted attention as the paradigm of gene activator proteins. It is now known that CRP, in conjunction with cAMP, participates as a global transcription factor in a wide regulatory network, both activating and repressing a large set of operons. One mechanism of catabolite repression is due to the reduction in the intracellular concentrations of both cAMP and CRP.
The CRP protein is composed of two identical subunits of 209 amino acid residues each. It is active when complexed with cAMP, which behaves as an allosteric effector. It is a prototype of sequence-specific DNA-binding proteins containing a helix–turn–helix motif. The cAMP–CRP complex binds to specific DNA sites in various operons. On binding DNA, it bends the DNA and interacts with RNA polymerase and/or other regulatory proteins to regulate transcription of the target operons.
Following early studies on the properties of purified CRP, the crp gene was cloned and sequenced in 1982. (3, 4) The X-ray crystallography structure of CRP complexed with cAMP was solved in 1981 (5) .This confirmed the dimeric nature and domain structure of CRP deduced from biochemical studies. The subunit has two domains (Fig. 1a). The larger N-terminal domain is responsible for cAMP binding and for dimerization. The smaller C-terminal domain contains a helix–turn–helix DNA-binding motif. CRP is one of the first regulatory proteins where the helix–turn–helix motif was identified. CRP alone is able to bind to DNA nonspecifically. The binding of cAMP induces a conformational state that binds to specific sequences with a dyad symmetry. The detailed nature of the structural changes in CRP caused by cAMP remains unknown.
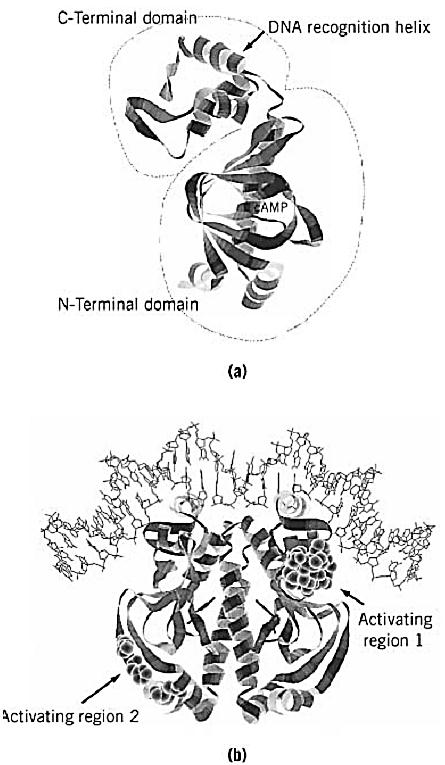
Figure 1. Structures of the CRP monomer and the CRP-DNA complex. The crystallographic coordinates were obtained from the Brookhaven Protein databank (accession code 1CGP). The images were generated by Protein Adviser (FQS). (a) The two domains of a CRP monomer. The larger N-terminal domain is responsible for cAMP binding and for dimerization. The smaller C-terminal domain contains a helix–turn–helix motif that is involved in DNA binding. (b) The CRP dimer bound to a consensus DNA site. The positions that are responsible for transcription activation (activating regions) are highlighted by space-filling models. Activating region 1 contacts the C-terminal domain of a subunit of RNA polymerase (8), while activating region 2 contacts the N-terminal domain of a subunit of RNA polymerase (9).
The DNA sequences of many CRP binding sites, and a resulting consensus sequence, have been determined. They include variations of the 22-bp palindromic sequence, 5′-AAATGTGATCTAGATCACATTT-3′, in which the two TGTGA motifs are relatively well conserved in different promoters. The binding of cAMP–CRP to target DNA induces bending of the DNA (6). The structure of cocrystals of cAMP-CRP bound to a 30-bp DNA target was determined in 1991. (7) It shows the helix-turn-helix motifs of the two subunits inserted into successive major groves of a DNA that is bent by 90°C (Fig. 1b). In promoters where CRP alone is sufficient to activate transcription, the role of the cAMP–CRP complex is to enhance functional binding of RNA polymerase (8, 9). Binding of cAMP–CRP or RNA polymerase to these promoters stimulates the other's binding. An example is the lac promoter, where the CRP-binding site is centered on base pair –61, which is one of the preferable positions for CRP action. Another position where CRP activates transcription efficiently is –41, as in the gal promoter. Changing these two standard distances reduces or eliminates CRP action. Only when this change is by an integral number of turns of DNA double helix is the ability of CRP to activate transcription retained to some extent, suggesting that both CRP and RNA polymerase must bind to the same face of the DNA (10, 11). Genetic and biochemical analysis revealed that cAMP–CRP activates transcription by directly contacting RNA polymerase at these simple promoters. A surface-exposed loop in the C-terminal domain of CRP and the C-terminal part of the a subunit of RNA polymerase are primarily responsible for the contact between two proteins (8) . An additional contact between the N-terminal domain of CRP and the N-terminal domain of a subunit is also involved in transcription activation at the promoters where the CRP site lies at –41 (9). The interaction between CRP and RNA polymerase is needed transiently to stimulate events leading to the formation of an open complex in these simple promoters (12). The contribution of CRP-induced DNA bending to transcription activation is still unclear. The CRP-binding site lies well upstream in several CRP-dependent promoters. In these cases, CRP acts as a coactivator of a second activator that is presumed to be responsible for the direct interaction with RNA polymerase. In the example of the araBAD promoter, CRP binds around –94 and the second activator AraC binds in the region between the CRP and RNA polymerase sites (13).
CRP also acts as a repressor or a corepressor in several operons. A simple example is the cya promoter, where cAMP–CRP inhibits the action of RNA polymerase by binding a target site located within the promoter (14). A more complex mechanism of repression is found in the operons that are coordinately regulated by CRP and CytR. Most of these promoters possess tandem CRP-binding sites that flank the CytR operator. An example is the deo promoter, where the CRP-binding sites are located at –41 and –94. The binding of cAMP-CRP to these sites dramatically enhances the binding of CytR to its operator, resulting in the formation of a repression complex containing both cAMP–CRP and CytR (15).
Although many studies have been identifying the regions of both transcription factors and RNA polymerase that are responsible for protein–protein and protein–DNA interactions, how these interactions lead to transcription activation is largely unknown. CRP, along with its target promoters, will continue to be a useful system for further understanding of molecular mechanisms of transcriptional regulation, including this fundamental question.
References
1. G. Zubay, D. Schwartz, and J. R. Beckwith (1970) Proc. Natl. Acad. Sci. USA 66, 104–110.
2. M. Emmer, B. deCrombrugghe, I. Pastan, and R. L. Perlman (1970) Proc. Natl. Acad. Sci. USA 66. 480–487 .
3. H. Aiba, S. Fujimoto, and N. Ozaki (1982) Nucleic Acids Res. 10, 1345–1361.
4. P. Cossart and B. Gicquel-Sanzey (1982) Nucleic Acids Res. 10, 1363–1378.
5. D. B. McKay and T. A. Steitz (1981) Nature 290, 744–749.
6. H.-M. Wu and D. M. Crothers (1984) Nature 308, 509–513.
7. S. C. Schultz, G. C. Shields, and T. A. Steitz (1991) Science 253, 1001–1007.
8. S. Busby and R. H. Ebright (1994) Cell 79, 743–746.
9. S. Busby and R. H. Ebright (1997) Mol. Microbiol. 23, 853–859.
10. K. Gaston, A. Bell, A. Kolb, H. Buc, and S. Busby (1990) Cell 62, 733–743.
11. C. Ushida and H. Aiba (1990) Nucleic Acids Res. 18, 6325–6330.
12. H. Tagami and H. Aiba (1998) EMBO J 17, 1759–1767.
13. R. B. Lobell and R. F. Schleif (1991) J. Mol. Biol. 218, 45–54.
14. H. Aiba (1985) J. Biol. Chem. 260, 3063–3070.
15. L. Søgaard-Anderson and P. Valentin-Hansen (1993) Cell 75, 557–566.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|