


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 10-6-2021
Date: 6-11-2020
Date: 18-5-2021
|
Base Pairs
DNA exists predominantly as an antiparallel double-stranded helix. The two strands are helically coiled, which maximizes the exposure of the negatively charged sugar–phosphate backbone to water and shields the hydrophobic aromatic bases in the middle from water. Figure 1 shows the Watson–Crick base pairs in which the specificity of base pairing is provided by hydrogen bonds. The complementary arrangements of hydrogen bond donors and acceptors allow the Watson–Crick base pairing of guanine with cytosine (G:C) and adenine with thymine (A:T). The two C1′ atoms within a G:C base pair and an A:T base pair are equidistant (~10.5 Å). It should be noted that other types of base pairs are also known to exist, and they often are involved in unusual DNA structures.
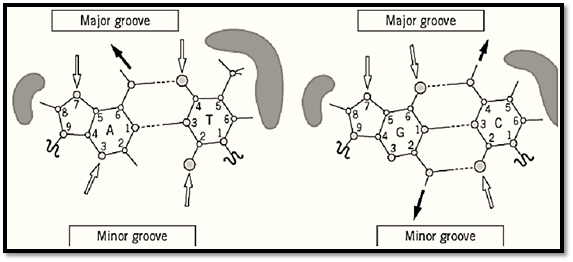
Figure 1. Schematic illustration of the A:T and G:C Watson–Crick base pairs. Hydrogen bonds are shown as dashed line Hydrogen bond donors (gray arrow) and acceptors (white arrow) of the bases are shown. Note that the bases can pair in more than one way. Hydrophobic edges of the base pairs are emphasized by gray shade. The major and minor groove edges of the base pair are shown.
Bases in a DNA double helix are stacked on top of each other. The stacking interaction is stabilized by the electronic p p interaction of the aromatic rings, and it also minimizes the exposure of the bases to the solvent. The base stacking has preferred interactions, depending on the characteristic dipole–dipole interactions between adjacent base pairs. Thus, the stacking patterns vary with the helix type and the particular base–base step. In B DNA, the 5′-purine-p-pyrimidine-3′ (Pu-Py) base-pair step has a good stacking overlap, and is thus more stable, whereas the 5′-pyrimidine-p-purine-3′ (Py-Pu) base-pair step has a poor stacking overlap, and is thus less stable (Figure 2). Such differences in stacking energy have strong implications in the bendability of DNA associated with protein binding. It has been noted that DNA sequences with Py-Pu steps in B-DNA are deformed more easily; therefore steps such as CpG, TpA, or TpG (=CpA) are often associated with kinked DNA structures.
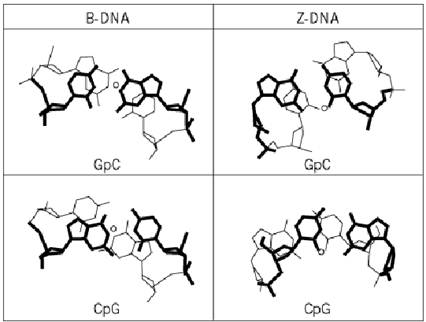
Figure 2. Stacking interactions in B-DNA (left) and Z-DNA (right) showing Pu-Py step stacking (top) and Py-Pu step stacking (bottom). The circle shows the position of the helix axis. Note that the Py-Pu steps (including CpG, TpA, and CpA (=TpG)) in B-DNA have very small stacking interactions, causing those steps to be easily deformed.
Through the high resolution X-ray structures of DNA oligonucleotides, it has been found that base pairs within a DNA duplex (or other structures) have significant departures from a strict coplanar geometry. The conformational parameters associated with the DNA base pairs are defined in Figure 3. For a more complete definition, the reader is referred to the Cambridge Convention (1).
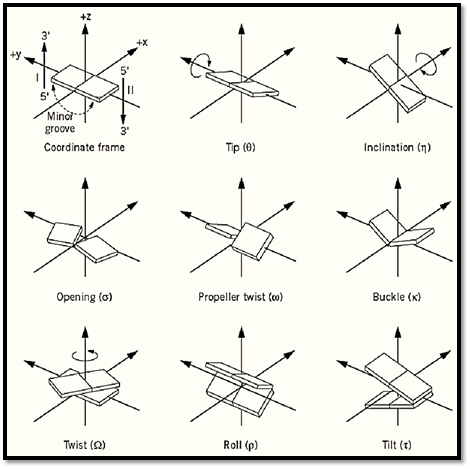
Figure 3. Definitions of various base pair conformations. (upper two rows) or two successive base pairs (bottom row.( In the top row the motions of the bases are coordinated, while in the middle row their motions are opposed. The left, center, and right columns describe rotations about the z, y, and x axes, respectively. The standard coordinate frame is defined at the upper left.
References
1. R. E. Dickerson et al. (1989) EMBO J. 8, 1–4.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|