


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 6-11-2020
Date: 21-11-2015
Date:
|
Aminoacyl tRNA Synthetases
The aminoacyl tRNA synthetases catalyze reactions that establish the rules of the genetic code. For this reason, there is great interest in these enzymes and their evolutionary development, which is thought to be closely connected to the establishment of the code. Research on the synthetases has led to the concept of an operational RNA code for amino acids that is imbedded in the acceptor stems of transfer RNA (tRNA) (1). The operational RNA code may have played an important role in the assembly of the genetic code and in the overall design of tRNA synthetases.
In the flow of genetic information, messenger RNA (mRNA) is transcribed from DNA, and the mRNA, in turn, is the template for protein synthesis (Fig. 1). The triplet codons of mRNA interact with the anticodons of tRNA through complementary base pairing. Amino acids joined to tRNA are incorporated into the growing polypeptide chain. The algorithm of the genetic code relates each amino acid to a specific trinucleotide codon. The triplet associated with a particular amino acid is determined in the aminoacylation reaction, where a given amino acid is linked to a tRNA bearing the anticodon trinucleotide that corresponds to that amino acid. These aminoacylation reactions are catalyzed by aminoacyl tRNA synthetases.
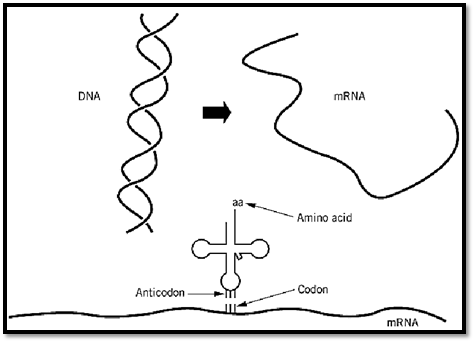
Figure 1. Flow of genetic information. Messenger RNA is synthesized from DNA. The mRNA has a string of trinucleotide codons that are translated into a polypeptide whose amino acid sequence is determined by the codons, according to the rules of the genetic code. The amino acid that is inserted into the polypeptide is determined by the codon–anticodon interaction with the tRNA that bears the amino acid corresponding to the particular anticodon. Therefore, the genetic code is determined by the linking of a particular amino acid with a particular anticodon triplet within a tRNA. The joining of amino acids to tRNA is catalyzed by aminoacyl tRNA synthetases. Thus, the genetic code is determined at the biochemical level in the aminoacylation reaction. (This figure was provided by Arturo Morales) .
1. Aminoacylation Reaction and the Genetic Code
For most tRNA synthetases, aminoacylation is carried out in a two-step reaction:
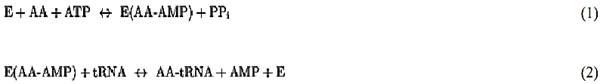
In the first reaction, the enzyme, E, uses ATP to activate an amino acid, AA, to yield the firmly bound aminoacyladenylate (AA-AMP). In the second step, the activated amino acid is transferred to the 3′-end of the tRNA, where it is connected by an ester linkage to the 2′- or 3′-hydroxyl group (after initial attachment, the amino acid can migrate back and forth between the 2′ and the 3′ positions). While most synthetases can carry out amino acid activation in the absence of tRNA, there are a few exceptions (such as glutaminyl-, glutamyl-, and argininyl-tRNA synthetases) that require the presence of the cognate tRNA for amino acid activation.
Because each of the 20 natural amino acids used in protein synthesis has a corresponding, or cognate, aminoacyl tRNA synthetase, there are 20 of these enzymes in each cellular compartment where proteins are synthesized. Each of them must distinguish its amino acid from all others and, at the same time, recognize the cognate tRNA that bears the anticodon corresponding to that amino acid. In prokaryotes and in the cytoplasm of eukaryotes, there is typically one tRNA synthetase for each amino acid (eukaryotic mitochondria have an additional set of synthetases that are essential for mitochondrial protein synthesis). However, the degeneracy of the genetic code means that there are 61 trinucleotides coding for the 20 amino acids. Reading these 61 triplets requires more than just 20 tRNAs. As a consequence, there are multiple tRNA isoacceptors for many of the synthetases. The synthetases for a particular amino acid must, therefore, recognize and aminoacylate all tRNA isoacceptors for that amino acid. This consideration in itself has important implications.
For example, the codons for serine are sixfold degenerate. In order to read these six codons, the serine tRNA isoacceptors must collectively permute all three anticodon nucleotides. Thus, for a single seryl-tRNA synthetase to aminoacylate all these tRNASer isoacceptors, the anticodon is not suitable for discrimination. Direct experiments in vitro and the X-ray crystallography structure of the seryl-tRNA synthetase-tRNASer complex have demonstrated that, in fact, seryl-tRNA synthetase does not contact the anticodon trinucleotide (2). This observation, and others described below, showed that, for at least some amino acids, the relationship between an amino acid and the triplet of the genetic code is not direct.
2. Classes of Aminoacyl tRNA Synthetases
The synthetases are heterogeneous in quaternary structures and subunit sizes, and this heterogeneity obscured more fundamental relationships between these enzymes. For example, in Escherichia coli, the quaternary structures of synthetases include a, a2, a4, and a2b2 (3). Subunit sizes vary from 303 to 951 amino acid residues (4). The 20 aminoacyl tRNA synthetases are now known to be divided into two classes of 10 enzymes each (Table 1) (8, 9). These classes are based on conserved sequence motifs and the structural architecture of the catalytic domains (8, 9). The classification is also based on the fact that the site of initial amino acid attachment on the tRNA differs between the two classes (8) .The classes appear to be fixed in evolution, because there is no example of an enzyme switching classes depending on the organism to which it belongs. Thus, the two classes may have developed early in evolution.
Table 1. Classes of Aminoacyl tRNA Synthetases
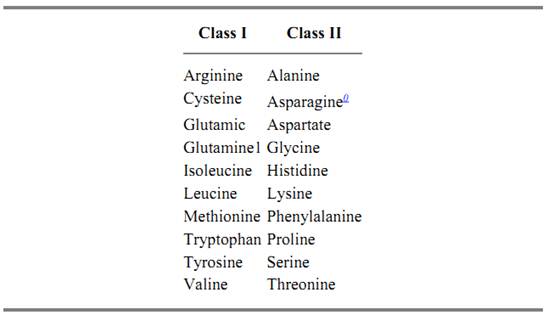
Gram-positive bacteria, plant chloroplasts, and animal mitochondria have been shown to have less than 20 tRNA synthetases. Instead, glutamyl-tRNA synthetase catalyzes attachment of glutamic acid to both tRNAGlu and tRNAGln and, similarly, aspartyl-tRNA synthetases catalyzes attachment of aspartate to both tRNAAsp and tRNAAsn. An amidotransferase then catalyzes the amidation of Glu-tRNAGln to give Gln-tRNAGln, and, likewise, amidation of Asp-tRNAAsn gives Asn-tRNAAsn (5-7).
2.1. Class I
These enzymes are usually monomers and are characterized by an architecture that is similar to that seen in dehydrogenases and other nucleotide-binding proteins. This structural motif is a Rossmann nucleotide-binding fold, which consists of alternating b-strands and a-helices (Fig. 2) (10-12). In the case of class I tRNA synthetases, the fold is divided into two b3a2 halves to give an overall b6a4 structure. In this structure, the b-strands are arranged in parallel. A polypeptide of variable length, designated as connective polypeptide 1 (CP1), links together the two halves of the active site (13). In some class I enzymes, this insertion plays a role in translational editing. It also contains some of the residues for binding the synthetase to the tRNA acceptor helix (14).
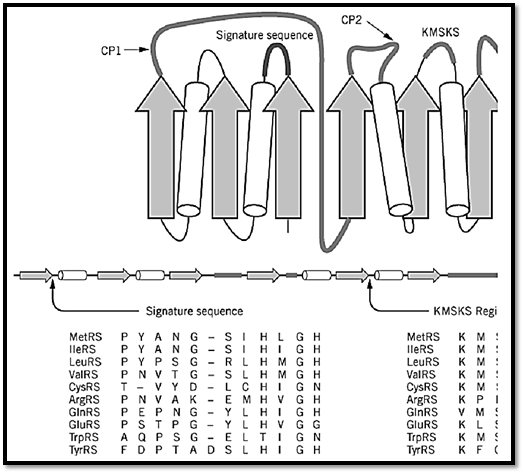
Figure 2. Design of a class I tRNA synthetase. The nucleotide-binding fold of class I tRNA synthetases consists of altern (cylinders) that form a b6a4 structure. A two-dimensional spatial arrangement of these elements is shown at the top, and abelow. A second domain of variable size occurs after the nucleotide-binding fold. The fold is split into two b3a2 halves bpolypeptide 1 (CPI). A second, smaller insertion (CP2) splits the second half of the fold. Two sequence elements were us enzymes. These are known as the 12-residue signature sequence, which ends in the HIGH tetrapeptides (10, 11) and as thlocations in the schematic structure are shown near the label signature sequence and KMSKS. By way of example, an ali sequences of the 10 class I E. coli enzymes is shown beneath the schematic figures. Similar alignments can be made for c throughout evolution. (This figure was provided by Arturo Morales.)
The nucleotide-binding fold contains the site for adenylate synthesis. This catalytic domain may be identified by two characteristic sequence motifs, without any knowledge of three-dimensional structure. One motif is the 11–amino acid element known as the signature sequence, which ends in the sequence–His–Ile–Gly–His, or HIGH in one-letter code (10, 11). This element is located in the first half of the nucleotide-binding fold at the end of the first b-strand and the beginning of the first a-helix. It was designated as a signature sequence because it served as a clear signature for a subgroup of related synthetases, before many crystal structures were determined. The second element is the KMSKS motif, located in the second half of the nucleotide-binding fold (15). These elements are critical parts of the active site.
2.2. Class II
The class II enzymes are mostly a2 dimers. The active sites of class II enzymes have a completely different architecture that harbors three characteristic sequence motifs. The structure consists of a seven-stranded antiparallel b-sheet with three a-helices (9, 16-18) (Fig. 3) (8, 9, 19). The three characteristic sequence motifs are known as motifs 1, 2, and 3. The sequences of these motifs are highly degenerate (8, 9). They consist of a helix–loop–strand, strand–loop–strand, and strand–helix, respectively. All three of these motifs form part of the active site.
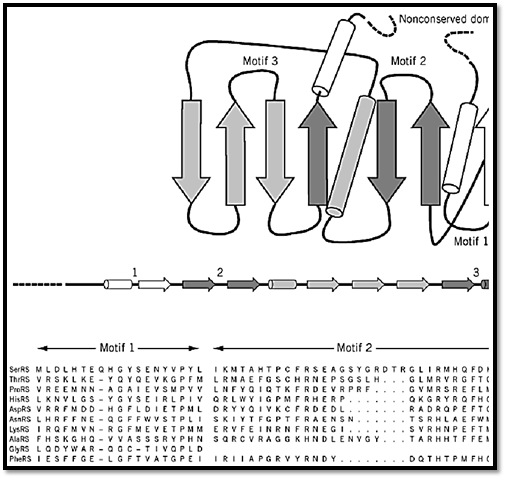
Figure 3. Design of a class II tRNA synthetase. The seven-stranded b-sheet with three a-helices is shown. The variable-s line that may occur on either the N- or the C-terminal side of the class-defining domain. Three characteristic sequence mand are distinguished in this illustration by their different shadings. These motifs are highly degenerate in sequence and c strand–loop–strand (motif 2), and strand–helix. The locations of these motifs in the class-defining domain are shown. An motifs for E. coli tRNA synthetases is also shown (8, 19). Note the high degeneracy of these sequence motifs, especially sequence elements in class I enzymes (Fig. 2). (This figure was provided by Dr. Arturo Morales).
2.3. Amino Acid Attachment
The site of initial amino acid attachment for class I enzymes is the 2′-hydroxyl, whereas the 3′-hydroxyl is used by class II enzymes (20). This distinction is now understood to result from a difference in the ways that the two enzymes approach the end of the tRNA. In particular, class I enzymes approach the end of the tRNA acceptor helix from the minor groove side, while class II enzymes approach from the major groove side (21).
3. Overall Structural Design and the Synthetase-tRNA Complex
The class-defining active-site domain is only a part of the tRNA synthetase structure. Inserted into the active-site domain are sequence motifs that enable the tRNA acceptor helix to bind with its 3′-end near the aminoacyl adenylate. These insertions are typically idiosyncratic to the synthetase. Two examples are the CP1 insertions of class I enzymes and the variable loops of motif 2 of class II enzymes, both of which have a role in docking the acceptor helices to the enzymes. However, in addition to insertions into the active-site domain, all synthetases have a second major domain. This domain, also typically idiosyncratic to the enzyme, provides for contacts with parts of the tRNA that are distal to the amino acid attachment site. For many (but not all) synthetases, this includes contacts with the anticodon. For the class I methionyl- and glutaminyl-tRNA synthetases, this second, anticodon-binding, domain is largely a-helical and largely b-structure, respectively (22, 23). This difference demonstrates that, even for enzymes in the same class, their second domains are completely unrelated. In the case of the class II seryl-tRNA synthetase, an unusual coiled-coil protrudes from the N-terminus of the enzyme. This structure, which is not found in many other class II enzymes, provides for contacts with the variable loop of tRNASer (2, 24).
Thus, to a rough approximation, the synthetases comprise two major domains (9, 14, 16, 23, 25-31). The tRNA molecule also comprises two major domains, which consist of the four arms of the cloverleaf secondary structure (Fig. 4). One domain is the acceptor-TyC minihelix, where the amino acid acceptor end and the TyC stem stack together to make a helix of 12 base pairs. The second domain is formed by stacking of the dihydrouridine stem with the anticodon stem. The result is an L-shaped three-dimensional structure where the amino acid acceptor end and the anticodon-containing template-reading-head are segregated into separate structural units (see Transfer RNA).
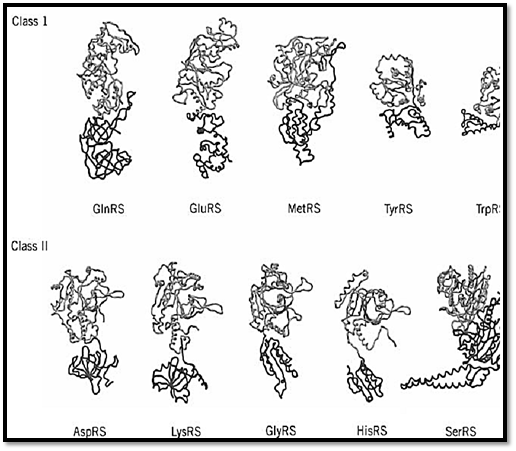
Figure 4. Examples of crystal structures of class I and class II tRNA synthetases. Regardless of the class to which an enz assigned, its structure can be approximated as comprising two major domains. One is the class-defining active site domain which is shared by all members of the same class. This domain (gray) is thought to be the historical tRNA synthetase. The second domain (dark gray) is typically idiosyncratic to the synthetase and is not shared by all members of the same class domain was probably added later to the synthetase structure. In these illustrations, the domains have been defined by the obvious visual divisions in the structures and, for that reason, the catalytic domain may extend somewhat beyond the regcontains the class-defining motifs. Some of these enzymes are homodimers; in all cases, only a single subunit is shown. I case of the tyrosyl tRNA synthetase, TyrRS, the structure of the second domain is not complete and therefore is truncated structural representation. Not shown is PheRS (25), which has an a2b2 quaternary structure. These structures were determ for GlnRS (14), GluRS (26), MetRS (23), TyrRS (27), TrpRS (28), AspRS (16), LysRS (29), GlyRS (30), HisRS (31), an SerRS (9). (This figure was provided by Arturo Morales).
The two major domains of the synthetase make contact with the two domains of the L-shaped tRNA molecule (Fig. 5). The catalytic domain, with insertions containing RNA-binding determinants, interacts with the acceptor-TyC minihelix. The second major domain interacts with the second domain of the tRNA, where interactions may extend as far as the anticodon trinucleotide or may involve contacts with special structures, such as the large variable loop of tRNASer. The details of the interactions between tRNA and synthetase are described in RNA-binding proteins.
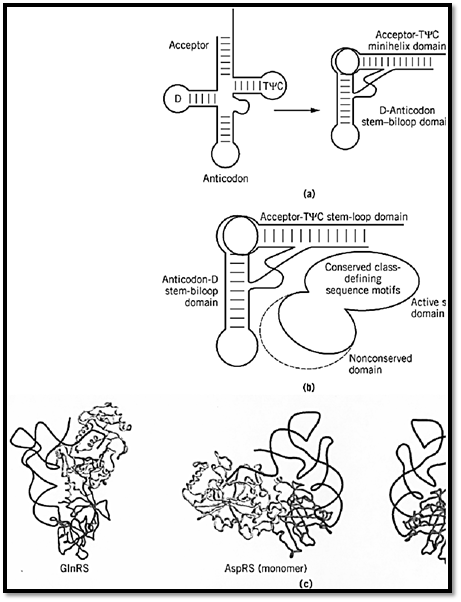
Figure 5. Two major domains of a synthetase interacting with two domains of a tRNA. (a) Schematic representation of the domains that segregate the amino acid attachment site into a 12-bp minihelix stem–loop and the anticodon triplet into a sTyC loops are indicated as common landmarks found in most all tRNA. (Adapted from Ref. 32.) (b) Domains of a syntheses of a tRNA. The second domain of the synthetase is shown with a dotted line to indicate that it varies in size and often ext33.) (c) Examples of synthetase-tRNA complexes, with the active-site class-defining domain interacting with the acceptor second, idiosyncratic domain of the synthetase interacting with the second domain of the tRNA (5, 16). Separate shades domains. In the case of aspartyl tRNA synthetase, the protein is a dimer. Binding of a single tRNA to the monomer is show domain interactions. The dimeric complex with two bound tRNA is also shown at the right.
4. Operational RNA Code for Amino Acids
In addition to seryl-tRNA synthetase, alanyl-tRNA synthetase is an example of a synthetase that makes no contact with the anticodon (34). Instead, an acceptor helix G3:U70 wobble base pair is a major determinant of the identity of an alanine tRNA (35, 36). Alteration of this base pair to G:C, A:U, I:U, or U:G abolishes aminoacylation with alanine (37-39). Transfer of this base pair into other, nonalanine tRNA confers alanine acceptance on them. Thus, the G3:U70 pair marks a tRNA for charging with alanine.
Because the G3:U70 base pair is located in the acceptor helix, further experiments tested whether the 12-bp acceptor-TyC minihelix by itself would be a substrate for aminoacylation (Fig. 6) (40-42). Not only the minihelix, but also a 7-base-pair(bp) microhelix consisting of just the acceptor stem, is efficiently charged with alanine, provided that it contains the G3:U70 base pair (40, 43). Transfer of this base pair into other microhelices confers alanine acceptance on them. Thus, the charging behavior of the mini- and microhelices reproduces that seen with the full tRNA.
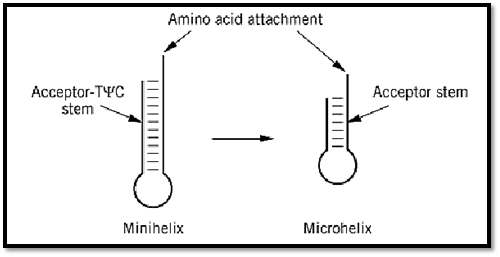
Figure 6. Minihelix and microhelix substrates for aminoacylation. The minihelix is derived from the 12-bp acceptor-TyC domain of the tRNA, while the microhelix is a hairpin helix whose base pairs consist of the 7-bp acceptor stem portion (40). These substrates for aminoacylation are devoid of the anticodon trinucleotides of the genetic code. About 11 examples of aminoacylation of minihelix or microhelix structures have been demonstrated. These substrates have been used to delineate the operational RNA code for amino acids (41). (Adapted from Ref. 42.)
The charging of specific RNA helices has now been demonstrated with at least 11 different tRNA synthetases, even for cases where the anticodon is known to play a significant role in the recognition of the related tRNA (41, 44-47). In the case of histidine, for example, an extra nucleotide at the 5′ end of the acceptor helix is characteristic of and unique to histidine tRNA throughout evolution. RNA microhelices that contain the extra base are charged with histidine (48). Thus, the 5′-appended nucleotide marks a molecule for charging with histidine. The smallest substrates seen to be charged with specific amino acids are stem–loop hairpins with as few as four base pairs stabilized by an RNA tetraloop motif (Fig. 7) (49).
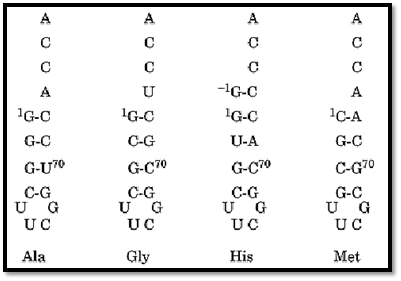
Figure 7. RNA tetraloop substrates for aminoacylation. The amino acid that can be charged onto the designated structures is indicated (49). These short helices are stabilized by an RNA tetraloop motif that confers unusual stability to short RNA helices. (Adapted from Ref. 49.)
The charging of microhelix substrates is sometimes considerably less efficient than that for the corresponding tRNA. In each case, however, charging is sequence-specific and depends on two to four nucleotides near the amino acid attachment site. In a fine-structure mapping of the efficiently charged alanine microhelix, a constellation of atoms was identified as critical for the aminoacylation signal. Prominent among these atoms was the exocyclic 2-amino group of G of the G3:U70 base pair. Also important were specific 2′-hydroxyl groups that fell within a 5-Å (Å = 10–10 m) radius of the critical 2-amino group (39).
These anticodon-independent aminoacylations of oligonucleotide substrates demonstrate that specific RNA sequences and structures per se, rather than the trinucleotides of the genetic code, correspond to specific amino acids. The relationship between these RNA sequences and structures and particular amino acids constitutes an operational RNA code for amino acids that is distinct from, but related to, the genetic code. This operational RNA code may have predated the genetic code (1).
5. Assembly of the Synthetase-tRNA Complex and Relationship of the Operational RNA Code to the Genetic Code
Several considerations led to the proposal that the minihelix and anticodon-containing second domain of tRNA had distinct origins (1, 50-52). The minihelix, with its amino acid attachment site, is viewed as the historical, or earliest, part of the tRNA. Similarly, the class-defining catalytic domain of tRNA synthetases is thought to be the historical enzyme, with the idiosyncratic second domain added later. Experiments with alanyl-tRNA synthetase have demonstrated that a relatively small piece of the enzyme (containing the active site) can by itself charge an RNA microhelix. This result directly demonstrated a domain–domain interaction between discrete units of the tRNA and the synthetase that may somewhat resemble an evolutionarily earlier system (33, 54).
Thus, the early synthetase may have consisted solely of a domain for adenylate synthesis. Insertions into this domain allowed the docking of RNA substrates near the activated amino acid so that aminoacylation could occur (Fig. 8). Addition of the second domain of the tRNA, with its anticodon-containing template reading head, and of the second domain of the synthetases was a second, later event. This event led to the joining of the operational RNA code to the genetic code. This scheme also suggests that the relationship between a particular amino acid and the triplet of the code is random, and simply depends on which anticodon-containing domain happened to be fused to the minihelix domain of the tRNA.
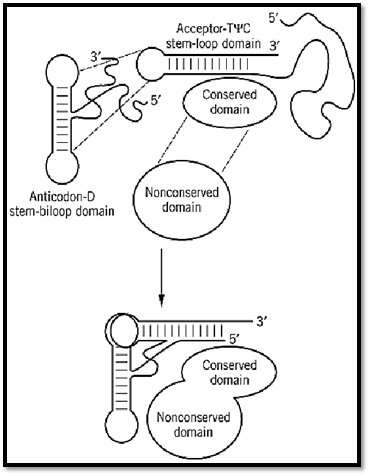
Figure 8. Assembly of a tRNA synthetase in evolution. A primordial tRNA synthetase is envisioned as interacting with a minihelix-like structure. As the tRNA structure developed, a template reading head (anticodon domain) was added, along with the second domain for the synthetase. (Adapted from Ref. 1.)
6. Conclusions
The tRNA synthetases may be among the earliest proteins, arising in evolution contemporaneously with the development of the genetic code. The first synthetases may have been ribozymes that catalyzed aminoacylation reactions with a specificity that depended on the sequences and structures of the RNA substrates (54, 55). The proteins that replaced these ribozymes were probably small. As a result, they could not extend much beyond the amino acid attachment site and, for that reason, gave rise to a system of interactions that based specificity of aminoacylation on interactions near the end of the acceptor helix. How these charged RNA substrates were used to synthesize specific proteins is a question of great interest.
References
1. P. Schimmel, R. Giegé, D. Moras, and S. Yokoyama (1993) Proc. Natl. Acad. Sci. USA 90 , 8763–8768 .
2. V. Biou, A. Yaremchuk, M. Tukalo, and S. Cusack (1994) Science 263, 1404–1410.
3. P. Schimmel (1987) Ann. Rev. Biochem. 56, 125–158.
4. S. A. Martinis and P. Schimmel (1996) in Escherichia coli and Salmonella (F. C. Neidhardt Jr., ed.), ASM Press, Washington, DC, pp. 887–901.
5. A. Schon, C. G. Kannangara, S. Gough, and D. Söll (1988) Nature 331, 187–190.
6. A. W. Curnow, M. Ibba, and D. Söll (1996) Nature 382, 589–590.
7. Y. Gagnon, L. Lacoste, N. Champagne, and J. Lapointe (1996) J. Biol. Chem. 271, 14856-14863 .
8. G. Eriani, M. Delarue, O. Poch, J. Gangloff, and D. Moras (1990) Nature 347, 203–206.
9. S. Cusack, C. Berthet-Colominas, M. Hartlein, N. Nassar, and R. Leberman (1990) Nature 347, 249–255 .
10. T. A. Webster, H. Tsai, M. Kula, G. A. Mackie, and P. Schimmel (1984) Science 226, 13151317- .
11. S. W. Ludmerer and P. Schimmel (1987) J. Biol. Chem. 262, 10801–10806.
12. C. Hountondji, F. Lederer, P. Dessen, and S. Blanquet (1986) Biochemistry 25, 16–21.
13. R. M. Starzyk, T. A. Webster, and P. Schimmel (1987) Science 237, 1614–1618.
14. M. A. Rould, J. J. Perona, D. Söll, and T. A. Steitz (1989) Science 246, 1135–1142.
15. C. Hountondji, P. Dessen, and S. Blanquet (1986) Biochimie 68, 1071.
16. M. Ruff, S. Krishnaswamy, M. Boeglin, A. Poterszman, A. Mitschler, A. Podjarny, B. Rees, J. C. Thierry, and D. Moras (1991) Science 252, 1682–1689.
17. S. Cusack, M. Hartlein, and R. Leberman (1991) Nucl. Acid Res. 19, 3489–3498.
18. D. Moras (1992) Trends Biochem. Sci. 17, 159–164.
19. S. Cusack (1993) Biochimie 75, 1077–1081.
20. P. R. Schimmel and D. Söll (1979) Annu. Rev. Biochem. 48, 601–648.
21. J. Cavarelli and D. Moras (1993) FASEB J. 7, 79–86.
22. M. A. Rould, J. J. Perona, D. Söll, and T. A. Steitz (1991) Nature 352, 213–218.
23. S. Brunie, C. Zelwer, and J. Risler (1990) J. Mol. Biol. 216, 411–424.
24. S. Cusack, A. Yaremchuk, and M. Tukalo (1996) EMBO J. 15, 2834–2842.
25. L. Mosyak, L. Reshetnikova, Y. Goldgur, M. Delarue, and M. G. Safro (1995) Nature Struct. Biol. 2, 537–547.
26. O. Nureki, D. G. Vassylyev, K. Katayanagi, T. Shimizu, S. Sekine, T. Kigawa, T. Miyazawa, S. Yokoyama, and K. Morikawa (1995) Science 267, 1958–1965.
27. P. Brick, T. N. Bhat, and D. M. Blow (1988) J. Mol. Biol. 208, 83–98.
28. S. Doublie, G. Bricogne, C. Gilmore, and C. W. Carter Jr. (1995) Structure 3, 17–31.
29. S. Onesti, A. D. Miller, and P. Brick (1995) Structure 3, 163–176.
30. D. T. Logan, M.-H. Mazauric, D. Kern, and D. Moras (1995) EMBO J. 14, 4156–4167.
31. J. G. Arnez, D. C. Harris, A. Mitschler, B. Rees, C. S. Francklyn, and D. Moras (1995) EMBO J. 14, 4143–4155.
32. J. J. Burbaum and P. Schimmel (1991) J. Biol. Chem. 266, 16965–16968.
33. D. D. Buechter and P. Schimmel (1993) Crit. Rev. Biochem. Mol. Biol. 28, 309–322.
34. S. J. Park and P. Schimmel (1988) J. Biol. Chem. 263, 16527–16530.
35. Y.-M. Hou and P. Schimmel (1988) Nature 333, 140–145.
36. W. H. McClain and K. Foss (1988) Science 240, 793–796.
37. S. J. Park, Y.-M. Hou, and P. Schimmel (1989) Biochemistry 28, 2740–2746.
38. K. Musier-Forsyth, N. Usman, S. Scaringe, J. Doudna, R. Green, and P. Schimmel (1991) Science 253, 784–786.
39. K. Musier-Forsyth and P. Schimmel (1992) Nature 357, 513–515.
40. C. Francklyn and P. Schimmel (1989) Nature 337, 478–481.
41. S. A. Martinis and P. Schimmel (1995) in tRNA: Structure, Biosynthesis and Function (D. Söll and U. L. RajBhandary, eds.), American Society for Microbiology, Washington, DC, pp. 349370- .
42. P. Schimmel (1993) in The Translation Apparatus (K. H. Nierhaus, F. Franceschi, A. R. Subramanian, V. A. Erdmann, and B. Wittmann-Liebold, eds.), Plenum Press, New York, pp. 13–21.
43. C. Francklyn, J.-P. Shi, and P. Schimmel (1992) Science 255, 1121–1125.
44. M. Frugier, C. Florentz, and R. Giegé (1994) EMBO J. 13, 2218–2226.
45. Y.-M. Hou, T. Sterner, and R. Bhalla (1995) RNA 1, 707–713.
46. C. L. Quinn, N. Tao, and P. Schimmel (1995) Biochemistry 34, 12489–12495.
47. M. E. Saks and J. R. Sampson (1996) EMBO J. 15, 2843–2849.
48. C. Francklyn and P. Schimmel (1990) Proc. Natl. Acad. Sci. USA 87, 8655–8659.
49. J.-P. Shi, S. A. Martinis, and P. Schimmel (1992) Biochemistry 31, 4931–4936.
50. A. M. Weiner and N. Maizels (1987) Proc. Natl. Acad. Sci. USA 84, 7383–7387.
51. H. F. Noller (1993) in The RNA World (R. F. Gesteland and J. F. Atkins, eds.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 137–156.
52. N. Maizels and A. Weiner (1994) Proc. Natl. Acad. Sci. USA 91, 6729–6734.
53. D. D. Buechter and P. Schimmel (1995) Biochemistry 34, 6014–6019; Correction, p. 16352.
54. J. A. Piccirilli, T. S. McConnell, A. J. Zaug, H. F. Noller, and T. R. Cech (1992) Science 256, 1420–1424
55. M. Illangasekare, G. Sanchez, T. Nickles, and M. Yarus (1995) Science 267, 643–647.



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
العتبة العباسية المقدسة تستعد لإطلاق الحفل المركزي لتخرج طلبة الجامعات العراقية
|
|
|