


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 23-3-2021
Date: 15-11-2020
Date: 3-12-2015
|
The Nucleosome Is the Subunit of All Chromatin
KEY CONCEPTS
-A nucleosome contains approximately 200 bp of DNA
and two copies of each core histone (H2A, H2B, H3, and
H4).
-DNA is wrapped around the outside surface of the protein octamer.
-The histone octamer has a structure of an H3 -H4 tetramer associated with two H2A-H2B dimers.
-Each histone is extensively interdigitated with its partner.
-All core histones have the structural motif of the histone fold. N- and C-terminal histone tails extend out of the nucleosome.
-H1 is associated with linker DNA and can lie at the point where DNA enters or exits the nucleosome.
The 10-nm particlesrepresent the fundamental building block of all chromatin, the nucleosome. The nucleosome contains about 200 bp of DNA associated with a histone octamer that consists of two copies each of histones H2A, H2B, H3, and H4. These are known as the core histones. FIGURE 1 illustrates their association and dimensions diagrammatically.

FIGURE 1. The nucleosome consists of approximately equal masses of DNA and histones (including H1). The predicted mass of a nucleosome that contains H1 is 262 kD.
The histones are small, basic proteins (rich in arginine and lysine residues), resulting in a high affinity for DNA. Histones H3 and H4 are among the most conserved proteins known, and the core histones are responsible for DNA packaging in all eukaryotes. H2A and H2B are also conserved among eukaryotes, but show appreciable species-specific variation in sequence, particularly in the histone tails. The core regions of the histones are even conserved in archaea and appear to play a similar role in compaction of archaeal DNA.
The shape of the nucleosome corresponds to a flat disk or cylinder of diameter 11 nm and height 6 nm. The length of the DNA is roughly twice the 34-nm circumference of the particle. The DNA follows a symmetrical path around the octamer. FIGURE 2 shows the DNA path diagrammatically as a helical coil that makes about one and two-thirds turns around the cylindrical octamer. Note that the DNA “enters” and “exits” on one side of the nucleosome.
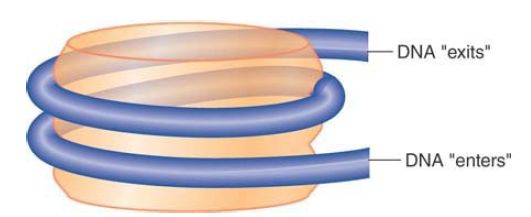
FIGURE 2 The nucleosome is a cylinder with DNA organized into ~one and two-thirds turns around the surface.
Viewing a cross section through the nucleosome in FIGURE 3, we see that the two circumferences made by the DNA lie close to each other. The height of the cylinder is 6 nm, of which 4 nm are occupied by the two turns of DNA (each of diameter 2 nm). The pattern of the two turns has a possible functional consequence.
One turn around the nucleosome takes about 80 bp of DNA, so 2 points separated by 80 bp in the free double helix can actually be close on the nucleosome surface, as illustrated in FIGURE 4.
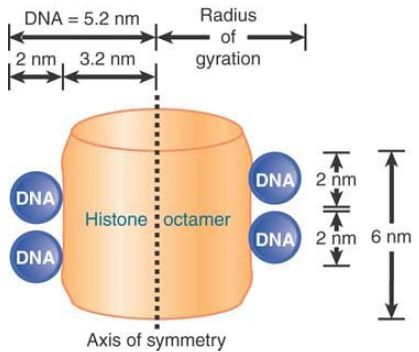
FIGURE 3 DNA occupies most of the outer surface of the nucleosome.
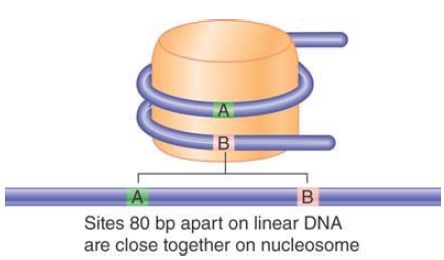
FIGURE 4 Sequences on the DNA that lie on different turns around the nucleosome may be close together.
The core histones tend to form two types of subcomplexes. H3 and H4 form a very stable tetramer in solution (H32 -H42 ). H2A and H2B most typically form a dimer (H2A-H2B). A space-filling model of the structure of the histone octamer (from the crystal structure at 3.1 Å resolution) is shown in FIGURE 5. Tracing the paths of the individual polypeptide backbones in the crystal structure shows that the histones are not organized as individual globular proteins, but that each is interdigitated with its partner: H3 with H4, and H2A with H2B. Figure 8.10 emphasizes the H32 -H42 tetramer (white) and the H2A-H2B dimer (blue) substructure of the nucleosome, but does not show individual histones.
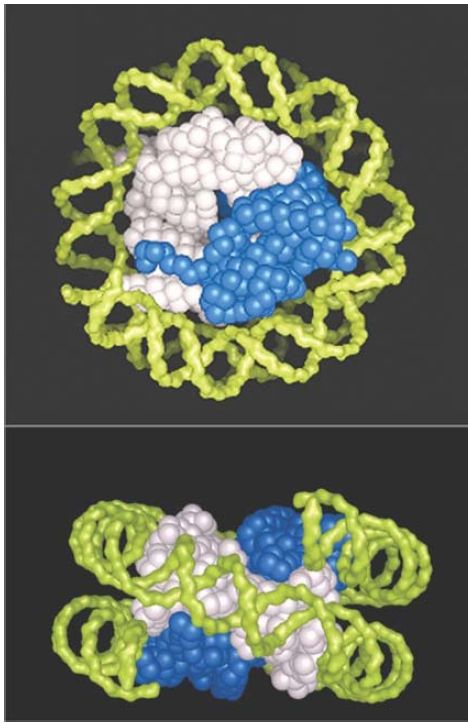
FIGURE 5 The crystal structure of the histone core octamer is represented in a space-filling model with the H32 -H42 tetramer shown in white and the H2A-H2B dimers shown in blue. Only one of the H2A-H2B dimers is visible in the top view, because the other is hidden underneath. The path of the DNA is modeled in green.
Photos courtesy of E. N. Moudrianakis, the Johns Hopkins University.
In the top view, you can see that the H32 -H42 tetramer accounts for the diameter of the octamer. It forms the shape of a horseshoe. The H3 -H4 tetramer alone can organize DNA in vitro into particles that display some of the properties of the core particle. The H2AH2B pairs fit in as two dimers, but you can see only one in this view. In the side view, we can distinguish the responsibilities of the H32 -H42 tetramer and of the separate H2A-H2B dimers. The protein forms a sort of spool, with a superhelical path that corresponds to the binding site for DNA, which is wound in about one and two-thirds turns in a nucleosome. The model displays twofold symmetry about an axis that would run perpendicular through the side view.
All four core histones show a similar type of structure in which three helices are connected by two loops. This highly conserved structure is called the histone fold, which you can see in FIGURE 6. These regions interact to form crescent-shaped heterodimers; each heterodimer binds 2.5 turns of the DNA double helix. Consistent with the need to package any DNA irrespective of sequence, binding is mostly to the phosphodiester backbone through a combination of salt links and hydrogen bonding interactions. In addition, an arginine side chain enters the minor groove of DNA at each of the 14 times it faces the octamer surface. FIGURE 7 shows a high-resolution view of the nucleosome (based on the crystal structure at 2.8 Å). The H32 -H42 tetramer is formed by interactions between the two H3 subunits, as you can see at the top of the nucleosome (in green) in the left panel of Figure 7. The association of the two H2A-H2B dimers on opposite faces of the nucleosome is visible in the right panel (in turquoise and yellow).
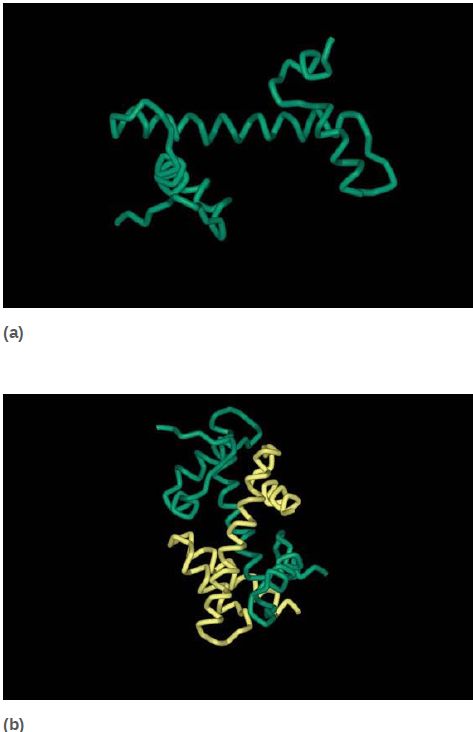
FIGURE 6 The histone fold (a) consists of two short α-helices flanking a longer α-helix. Histone pairs (H3 + H4 and H2A + H2B) interact to form histone dimers (b).
Data from: Arents, G., et al. 1991. “Structures from Protein Data Bank 1HIO.” Proc Natl
Acad Sci USA 88:10145–10152.
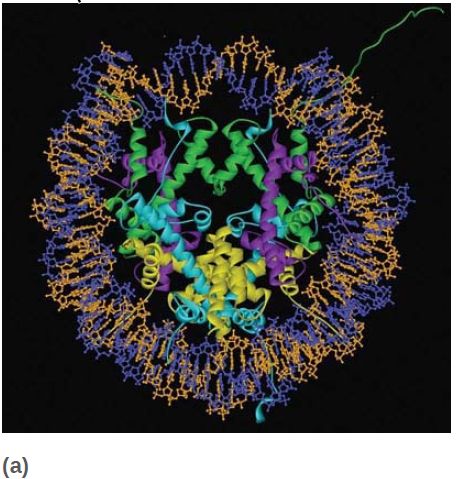
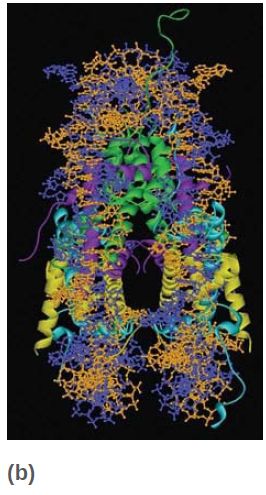
FIGURE 7 The crystal structure of the histone core octamer is represented in a ribbon model, including the 146-bp DNA phosphodiester backbones (orange and blue) and eight histone protein main chains (green: H3; purple: H4; turquoise: H2A; yellow: H2B).
Data from: Luger, K., et al. 1997. “Structures from Protein Data Bank 1AOI.” Nature 389:251–260.
Each of the core histones has a histone fold domain that contributes to the central protein mass of the nucleosome, sometimes referred to as the globular core. Each histone also has a flexible N-terminal tail (H2A and H2B have C-terminal tails, as well), which contains sites for covalent modification that are important in chromatin function. The tails, which account for about one-quarter of the protein mass, are too flexible to be visualized by X-ray crystallography; therefore, their positions in the nucleosome are not well defined, and they are generally depicted schematically, as shown in FIGURE 8. However, the points at which the tails exit the nucleosome core are known, and we can see the tails of both H3 and H2B passing between the turns of the DNA super-helix and extending out of the nucleosome, as shown in FIGURE 9.
The tails of H4 and H2A extend from both faces of the nucleosome. When histone tails are crosslinked to DNA by UV irradiation, more products are obtained with nucleosomes compared to core particles, which could mean that the tails contact the linker DNA. The tail of H4 is able to contact an H2A-H2B dimer in an adjacent nucleosome, which might contribute to the formation of higher-order structures .
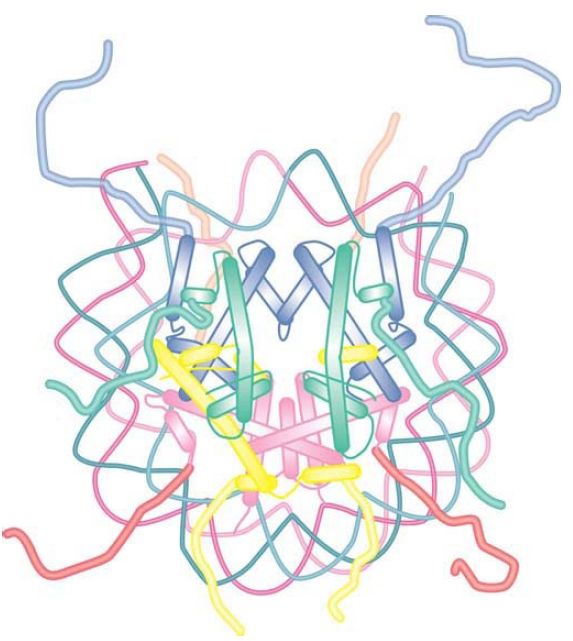
FIGURE 8 The histone fold domains of the histones are located in the core of the nucleosome. The N- and C-terminal tails, which carry many sites for modification, are flexible and their positions cannot be determined by crystallography.
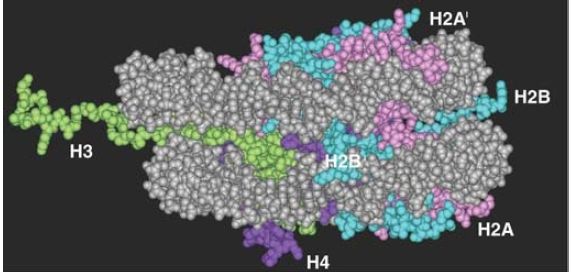
FIGURE 9 The histone tails are disordered and exit from both faces of the nucleosome and between turns of the DNA. Note this figure shows only the first few amino acids of the tails, because the complete tails were not present in the crystal structure.
Data from: Luger, K., et al. 1997. “Structure from Protein Data Bank 1AOI.” Nature 389:251–260.
The linker histones also play an important role in the formation of higher-order chromatin structures. The linker histone family, typified by histone H1, comprises a set of closely related proteins that show appreciable variation among tissues and among species. The role of H1 is different from that of the core histones. H1 can be removed without affecting the structure of the nucleosome, consistent with a location external to the particle, and only a subset of nucleosomes is associated with linker histones in vivo.
Nucleosomes that contain linker histones are sometimes referred to as chromatosomes. The precise interaction of histone H1 with the nucleosome is somewhat controversial. H1 is retained on nucleosome monomers that have at least 165 bp of DNA, but does not bind to the 146-bp core particle. The binding of H1 to a nucleosome also facilitates the wrapping of two full turns of DNA. This is consistent with the localization of H1 in the region of the linker DNA immediately adjacent to the core DNA. Although the precise positioning of linker histones remains somewhat controversial, protein crosslinking and structural studies are consistent with a model whereby H1 interacts with either the entry or exit DNA in addition to the central turn of DNA on the nucleosome, as shown in FIGURE 10. In this position, H1 has the potential to influence the angle of DNA entry or exit, which might contribute to the formation of higher-order structures .
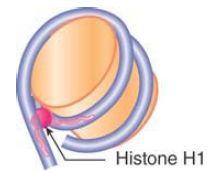
FIGURE 10 Possible model for the interaction of histone H1 with the nucleosome. H1 can interact with the central gyre of the DNA at the dyad axis, as well as with the linker DNA at either the entry or exit.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|