


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 15-11-2020
Date: 14-12-2015
Date: 10-3-2021
|
Beta-Lactoglobulin
b-Lactoglobulin is the major whey protein in the milk of ruminants, and it has also been reported in milk from many other species, although not human, lagomorph, or rodent. Where found, genetic variants have also been reported in most cases. Isolation of whey protein from bovine milk is straightforward (1), and its concentration is 2 to 3 g/L. Consequently, the protein has been used since its first isolation in 1934 as a convenient, small, soluble protein with which to develop and calibrate new techniques. For example, it is used as a component of standard mixtures for isoelectric focusing and SDS-PAGE and a calibration sequence for automatic sequenators (see Protein Sequencing). The extensive literature on b-lactoglobulin, largely that from the domestic cow, can be broadly divided into studies related to the molecule as a protein and those related to its importance to the dairy industry. Reviews of both abound (2-6).
The polypeptide chain of the protein contains about 160 residues. That of ruminant species is dimeric, and in other species it is monomeric. Figure 1 shows some of the known sequences from the SWISSPROT database (7). Disulfide bonds link cysteines 66 to 160 and 106 to 119. The asterisks show structurally conserved regions that were recognized following the structural determination first of the homologous plasma retinol-binding protein and then of bovine b-lactoglobulin (8, 9). These motifs define a widely distributed family called the lipocalins (10, 11), whose structure is an eight-stranded antiparallel b-barrel with (+1)8 topology (see Beta-Sheet) and with an a-helix on the outer surface. The structure of b-lactoglobulin is shown in Fig. 2 (12). The family relationship has also been observed in the gene sequences, which show similar arrangements of introns and exons (13). The 400 bp upstream of the open reading frame controls the high level of expression and its restriction to the mammary gland (14). This has led to successful use of the b-lactoglobulin operon in transgenic technology (15).

Figure 1. Sequences of mature b-lactoglobulins with the totally conserved residues in bold. The sites of possible genetic shown in the bovine sequence in italics, and changes in the bovine B variant are shown above. The N-terminal sequence (34). The asterisks show the structurally conserved regions indicative of the lipocalin family.
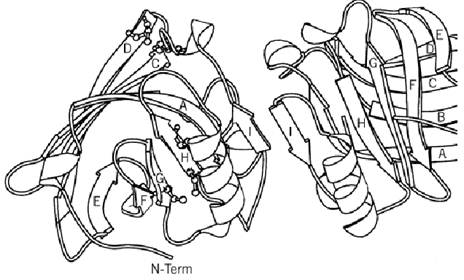
Figure 2. A MOLSCRIPT (35) diagram of the dimer of b-lactoglobulin showing the fold of the main chain.
No definite function has yet been ascribed to b-lactoglobulin, but its presence in a family containing mostly small, secreted proteins that transport hydrophobic molecules perhaps implies that its function, other than the obvious nutritional one, is associated with the transport or uptake of fatty acids or retinol, both of which it binds (16, 17). Indeed, over 20 different ligands have been reported that have association constants varying between 5 × 107 and 4 × 102M (3). By analogy with other lipocalins, the binding site should be in the central pocket formed by the b-strands, but there is no definitive evidence that this is the case. Recently it has been shown that palmitate and retinol can bind independently and simultaneously (18).
Some physical parameters of the protein are summarized in Table 1. The bovine protein is stable down to pH 2, and conformational studies have shown that there are several pH-dependent changes between pH 2 and pH 9 (19, 20). At the extremes of pH, the dimer dissociates, and it aggregates above pH 8.5 to 9.0. The conformational change between pH 6 and 8 leads to increased reactivity of the free cysteine residue, perhaps triggered by the titration of a carboxyl group with a pKa of 7.3. An association of four dimers forms an octamer at 5°C and pH 4.5 mainly in the A variant. This probably results from a carboxyl–carboxylate interaction involving the Gly64Asp change between the most common B and A genetic variants of b-lactoglobulin (21).


The complex behavior during refolding from urea between 2° and 25°C is ascribed to disulfide interchange. Cys106 pairs with either Cys119 or Cys121, and the former is the native (22, 23). It is not yet clear whether this also explains the partially folded form observed at pH 2, where the protein is predominantly a monomer (24). Refolding from guanidinium chloride at low pH shows a transient increase in helical content (25), an effect that is stabilized in ethanolic solution (26).
b-Lactoglobulin has been cloned and expressed both in Escherichia coli and in yeast, allowing creation of site-directed mutations (27) for probing the solution properties (28) or modifying the thermal denaturing behavior (29). Thermal denaturation is of particular interest to the dairy industry because heating milk is the basis of much processing and also the genetic variants exhibit different behavior (30). In addition, protein concentration, pH, temperature, ionic strength, dielectric constant, and buffer type all contribute to the denaturing behavior. Differential scanning calorimetry and thermal aggregation studies show that the A variant is less stable than the B variant (31), but studies at lower protein concentration show that the reverse is true (32). Thermal aggregation also involves intermolecular disulfide interchange, presumably initiated by the free thiol group (33).
References
1.R. Aschaffenburg and R. Drewry (1957) Biochem. J. 65, 273–277.
2.J. M. A. Tilley (1960) Dairy Sci. Abstr. 22, 111–125.
3.R. L. J. Lyster (1972) J. Dairy Res. 39, 279–318.
4.S. G. Hambling, A. S. McAlpine, and L. Sawyer (1992) In Advanced Dairy Chemistry I. (P. F. Fox, ed.), Elsevier, Amsterdam, pp. 141–190.
5.M. McSwiney, H. Singh, O. Campanella, and L. K. Creamer (1994) J. Dairy Res. 61, 221–232.
6.J. E. Kinsella and D. M. Whitehead (1987) Adv. Food Nutr. Res. 33, 343–438.
7.A. Bairoch and B. Boeckmann (1994) Nucleic Acid Res. 22, 3578-
8.M. Newcomer et al. (1984) EMBO J. 3, 1451–1454.
9.M. Z. Papiz et al. (1986) Nature 324, 383–385.
10.S. Pervaiz and K. Brew (1987) FASEB J. 1, 209–214.
11.D. R. Flower (1996) Biochem. J. 318, 1–14.
12.S. Brownlow et al. (1997) Structure, 5, 481–495.
13.S. Ali and A. J. Clark (1988) J. Mol. Biol. 189, 415–426.
14.J. P. Simons, M. McClenaghan, and A. J. Clark (1987) Nature 328, 530–532.
15.A. L. Archibald et al. (1990) Proc. Natl. Acad. Sci. USA 87, 5178–5182.
16.A. A. Spector and J. E. Fletcher (1970) Lipids 5, 403–411.
17.F. D. Fugate and P. S. Song (1980) Biochim. Biophys. Acta 652, 28–42.
18.M. Narayan and L. J. Berliner (1997) Protein Sci. 7, 150–157.
19. C. Tanford, L. G. Bunville, and Y. Nozaki (1959) J. Am. Chem. Soc. 81, 4032–4036.
20.S. N. Timasheff, L. Mescanti, J. J. Basch, and R. Townend (1966) J. Biol. Chem. 241, 2496–2501.
21.J. Witz, S. N. Timasheff, and V. Luzzati (1964) J. Am. Chem. Soc. 86, 168–173.
22. H. A. McKenzie, G. B. Ralston, and D. C. Shaw (1972) Biochemistry 11, 4539–4547.
23.T. E. Creighton (1980) J. Mol. Biol. 137, 61–80.
24.L. Ragona et al. (1997) Folding and Design 2, 281–290.
25.D. Hamada, S. Segawa, and Y. Goto (1996) Nature Struct. Biol. 3, 868–873.
26.E. Dufour, H. C. Bertrand, and T. Haertlé (1993) Biopolymers 33, 589–598.
27.C. A. Batt, L. D. Rabson, D. W. S. Wong, and J. E. Kinsella (1990) Agric. Biol. Chem. 54, 949–955.
28.Y. Katakura, M. Totsuka, A. Ametani, and S. Kaminogawa (1994) Biochim. Biophys. Acta .1207,58-67.
29.C. A. Batt, J. Brady, and L. Sawyer (1994) Tr. Food Sci. 5, 261–265.
30.E. Jakob and Z. Puhan (1992) Int. Dairy J. 2, 157–178.
31.X. L. Huang, G. L. Catignani, and H. E. Swaisgood (1994) J. Agric. Food Chem. 42, 1276–1280.
32.G. I. Imafadon, K. F. Ng-Kwai-Hang, V. R. Harwalkar, and C.-Y. Ma (1991) J. Dairy Res. 74, 2416-2422.
33.S. P. F. M. Roefs and K. G. Dekruif (1994) Eur. J. Biochem. 226, 883–889.
34.N. Azuma and K. Yamauchi (1991) Comp. Biochem. Physiol. 99B, 917–921.
35. P. J. Kraulis (1991) J. Appl. Cryst. 24, 946–950.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|