


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 15-5-2016
Date: 30-11-2015
Date: 28-12-2015
|
Chromosome Condensation Is Caused by Condensins
KEY CONCEPTS
- SMC proteins are ATPases that include condensins and cohesins.
- A heterodimer of SMC proteins associates with other subunits.
- Condensins cause chromatin to be more tightly coiled by introducing positive supercoils into DNA.
- Condensins are responsible for condensing chromosomes at mitosis.
- Chromosome-specific condensins are responsible for condensing inactive X chromosomes in C. elegans.
The structures of entire chromosomes are influenced by interactions with proteins of the structural maintenance of chromosome (SMC) family. These are ATPases that fall into two functional groups: condensins and cohesins. Condensins are involved in the control of overall structure and are responsible for the condensation into compact chromosomes at mitosis. Cohesins play a role in the connections between sister chromatids that concatenate through a cohesin ring, which must be released at mitosis. Both consist of dimers formed by SMC proteins.
Condensins form complexes that have a core of the heterodimer SMC2–SMC4 associated with other (non-SMC) proteins. Cohesins have a similar organization but consist of SMC1 and SMC3 and also interact with smaller non-SMC subunits, Scc1/Rad21 and Scc3/SA.
FIGURE 1shows that an SMC protein has a coiled-coil structure in its center that is interrupted by a flexible hinge region. Both the amino and carboxyl termini have ATP- and DNA-binding motifs. The ATP-binding motif is also known as a Walker module. SMC monomers fold at the hinge region, forming an antiparallel interaction between the two halves of each coiled coil. This allows the amino and carboxyl termini to interact to form a “head” domain. Different models have been proposed for the actions of these proteins depending on whether they dimerize by intra- or intermolecular interactions.
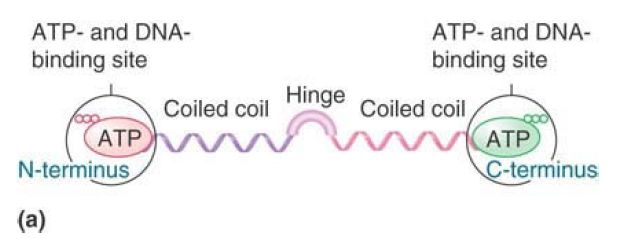
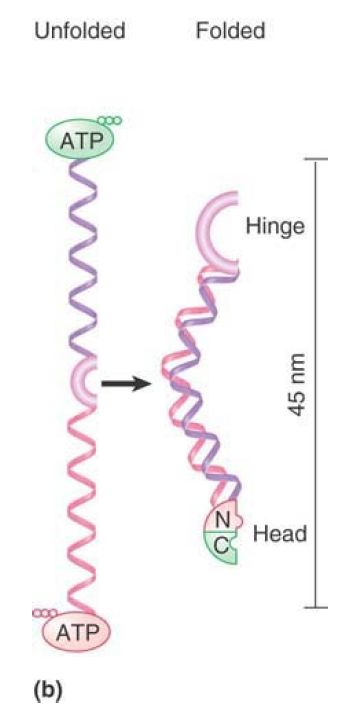
FIGURE 1. (a) An SMC protein has a Walker module with an ATP-binding motif and DNA-binding site at each end, which are connected by coiled coils that are linked by a hinge region. (b) SMC monomers fold at the hinge regions and interact along the length of the coiled coils. The N- and C-termini interact to form a head domain.
Data from I. Onn, et al., Annu. Rev. Cell Dev. Biol. 24 (2008): 105–129.
Folded SMC proteins form dimers via several different interactions. The most stable association occurs between hydrophobic domains in the hinge regions. FIGURE 2 shows that these hinge–hinge interactions result in V-shaped structures. Electron microscopy shows that in solution cohesins tend to form Vs, with the arms separated by a large angle, whereas condensins form more linear structures, with only a small angle between the arms. In addition, the heads of the two monomers can interact, closing the V, and the coils of the individual monomers may also interact with each other. Various non-SMC proteins interact with SMC dimers and can influence the final structure of the dimer.
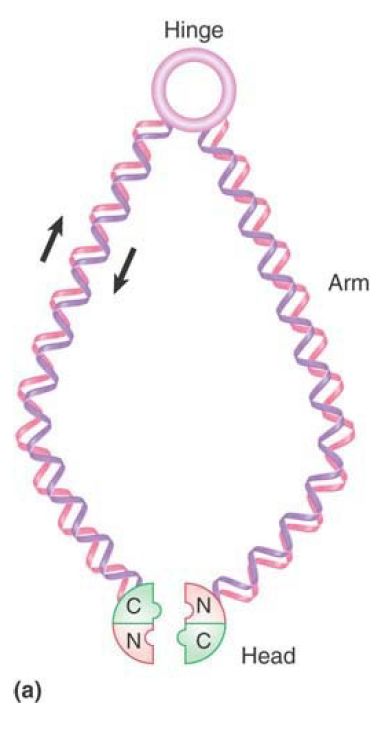
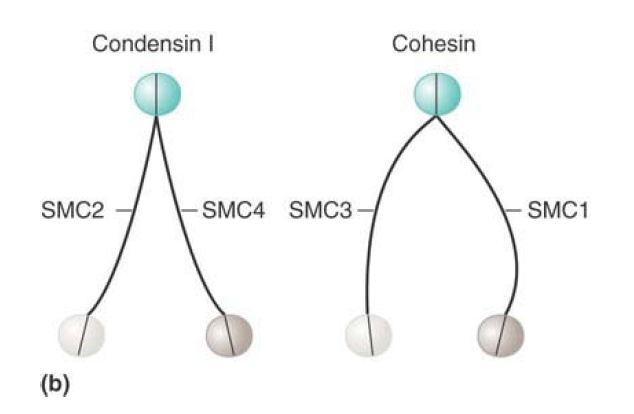
FIGURE 2. (a) The basic architecture of condensin and cohesin complexes. (b) Condensin and cohesin consist of V-shaped dimers of two SMC proteins interacting through their hinge domains. The two monomers in a condensin dimer tend to exhibit a very small separation between the two arms of the V; cohesins have a much larger angle of separation between the arms.
Data from T. Hirano, Nat. Rev. Mol. Cell Biol. 7 (2006): 311–322.
The function of cohesins is to hold sister chromatids together, but it is not yet clear how this is achieved. Several different models have been proposed for cohesin function. FIGURE 3 shows one model in which a cohesin could take the form of extended dimers, interacting hinge to hinge, that crosslink two DNA molecules. Head–head interactions would create tetrameric structures, adding to the stability of cohesion. An alternative “ring” model is shown in FIGURE 4. In this model, dimers interact at both their head and hinge regions to form a circular structure. Instead of binding directly to DNA, a structure of this type could hold DNA molecules together by encircling them.
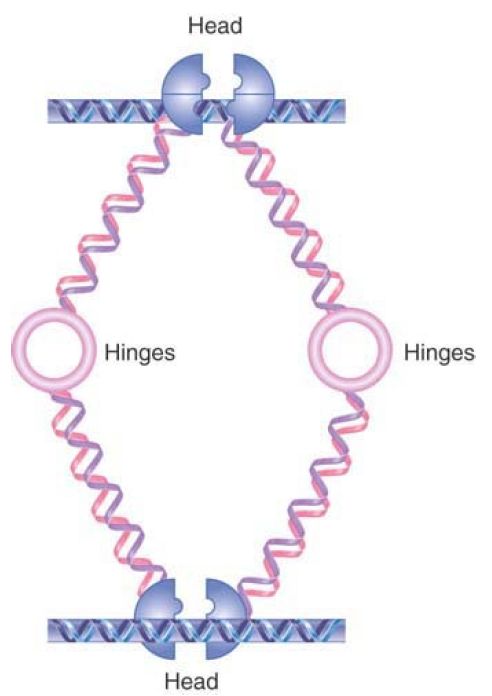
FIGURE 3. One model for DNA linking by cohesins. Cohesins may form an extended structure in which each monomer binds DNA and connects via the hinge region, allowing two different DNA molecules to be linked. Head domain interactions can result in binding by two cohesin dimers.
Data from I. Onn, et al., Annu. Rev. Cell Dev. Biol. 24 (2008): 105–129.
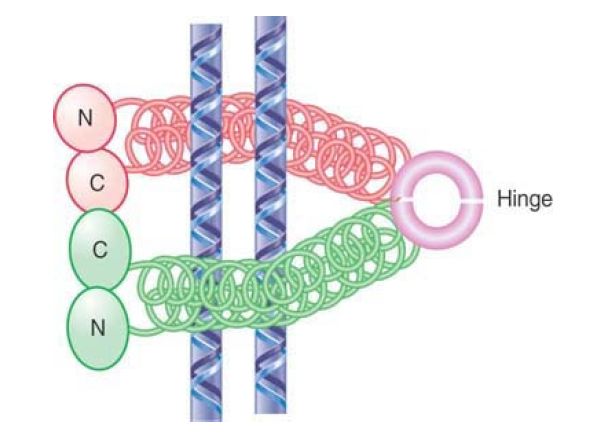
FIGURE 4. Cohesins may dimerize by intramolecular connections and then form multimers that are connected at the heads and at the hinge. Such a structure could hold two molecules of DNA together by surrounding them.
Whereas cohesins act to hold separate sister chromatids together, condensins are responsible for chromatin condensation. FIGURE 5 shows that a condensin could take the form of a V-shaped dimer, interacting via the hinge domains, that pulls together distant sites on the same DNA molecule, causing it to condense. It is thought that dynamic head–head interactions could act to promote the ordered assembly of condensed loops, but the details of condensin action are still far from clear.
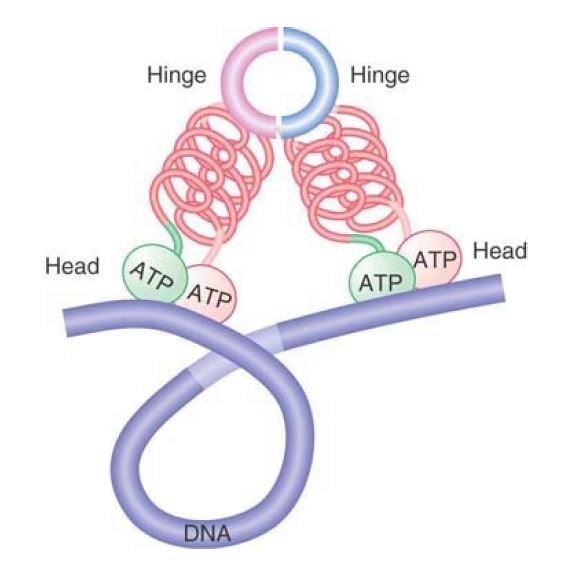
FIGURE 5. Condensins may form a compact structure by bending at the hinge, causing DNA to become compacted.
Visualization of mitotic chromosomes shows that condensins are located all along the length of the chromosome, as shown in FIGURE 6 . (By contrast, cohesins are found at discrete locations in a focal nonrandom pattern with an average spacing of about 10 kb.) The condensin complex was named for its ability to cause chromatin to condense in vitro. It has an ability to introduce positive supercoils into DNA in an action that uses hydrolysis of ATP and depends on the presence of topoisomerase I. This ability is controlled by the phosphorylation of the non-SMC subunits, which occurs at mitosis. It is not yet known how this connects with other modifications of chromatin—for example, the phosphorylation of histones. The activation of the condensin complex specifically at mitosis makes it questionable whether it is also involved in the formation of interphase heterochromatin. Recent evidence indicates that chromosome condensation does not involve hierarchal folding of chromatin into scaffolds but rather that the condensation process is dynamic. This dynamic process involves interactions of condensin between segments of chromatin that can be quite some distance apart. Therefore, chromosome condensation may involve a scaffold-free organization that consists of nucleosome fibers folded in an irregular manner in a polymer structure.
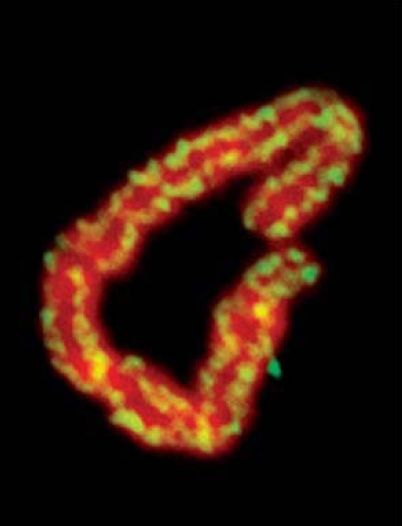
FIGURE 6. Condensins are located along the entire length of a mitotic chromosome. DNA is red; condensins are yellow.
Photo courtesy of Ana Losada and Tatsuya Hirano.
As discussed in the previous section, dramatic chromosomal changes occur during X-inactivation in female mammals and in X chromosome upregulation in male flies. In the nematode C.elegans, a third approach is used: twofold reduction of Xchromosome transcription in XX hermaphrodites relative to XO males. A dosage compensation complex (DCC) is maternally provided to both XX and XO embryos, but it then associates with both X chromosomes only in XX animals, while remaining diffusely distributed in the nuclei of XO animals. The protein complex contains an SMC core and is similar to the condensin complexes
that are associated with mitotic chromosomes in other species. This suggests that it has a structural role in causing the chromosome to take up a more condensed, inactive state. Recent studies have shown, though, that SMC-related proteins may also have roles in dosage compensation in mammals: The protein SmcHD1 (SMC-hinge domain 1) may actually contribute to the deposition of DNA methylation on the mammalian inactive X chromosome. SMCs could recruit DNA methyltransferase via a component of the SMC core that is involved in RNAi-directed DNA methylation, such as occurs in Arabidopsis via the DMS3 protein (another SMC-related protein).
Whatever the mechanism of transcriptional downregulation, multiple sites on the X chromosome appear to be needed for the DCC to be fully distributed along it, and short DNA sequence motifs have been identified that appear to be key for localization of DCC. The complex binds to these sites and then spreads along the chromosome to cover it more thoroughly.
Changes affecting all the genes on a chromosome, either negatively (mammals and C. elegans) or positively (Drosophila), are therefore a common feature of dosage compensation. The components of the dosage compensation apparatus may vary, however, as well as the means by which it is localized to the chromosome. Dosage compensation in mammals and Drosophila both entail chromosome-wide changes in histone acetylation and involve noncoding RNAs that play central roles in targeting X chromosomes for global change. In C. elegans, chromosome condensation by condensin homologs is used to accomplish dosage compensation. It remains to be seen whether there are also global changes in histone acetylation or other modifications in XX C. elegans that reflect the twofold reduction in transcription of the X chromosomes.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|