


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-4-2021
Date: 20-5-2021
Date: 16-11-2020
|
Termination Codons Are Recognized by Protein Factors
KEY CONCEPTS
- Termination codons are recognized by protein release factors, not by aminoacyl-tRNAs.
- The structures of the class 1 release factors (RF1 and RF2 in E. coli) resemble aminoacyl-tRNA–EF-Tu and EFG.
- The class 1 release factors respond to specific termination codons and hydrolyze the polypeptide–tRNA linkage.
- The class 1 release factors are assisted by class 2 release factors (such as RF3) that depend on GTP.
- The mechanism of termination in bacteria (which have two types of class 1 release factors) is similar to that of eukaryotes (which have only one class 1 release factor).
Two stages are involved in ending translation. The termination reaction itself involves release of the polypeptide chain from the last tRNA. The posttermination reaction involves release of the tRNA and mRNA and dissociation of the ribosome into its subunits.
None of the termination codons normally have tRNAs that can pair with them. They function in an entirely different manner from other codons and are recognized directly by protein factors. (The reaction does not depend on codon–anticodon recognition, so there seems to be no particular reason why it should require a triplet sequence. Presumably this is an evolutionary consequence of the genetic code.)
Termination codons are recognized by class 1 release factors (RFs). In E. coli, two class 1 release factors are specific for different codons. RF1 recognizes UAA and UAG, and RF2 recognizes UGA and UAA. The factors act at the ribosomal A site and require polypeptidyl-tRNA in the P site. The reading frames are present at much lower levels than initiation or elongation factors, with about 600 molecules of each per cell, equivalent to one reading frame per 10 ribosomes. At one time there was probably only a single release factor that recognized all termination codons, which later evolved into two factors with specificities for particular codons. Eukaryotes have a single class 1 release factor, eRF. The efficiency with which the bacterial factors recognize their target codons is influenced by the bases on the 3′ side.
The class 1 release factors are assisted by class 2 release factors, which are not codon specific. The class 2 factors are GTPbinding proteins. In E. coli, the role of the class 2 factor, RF3, is to release the class 1 factor from the ribosome. RF3 is a GTP-binding protein that is related to the elongation factors.
Although the general mechanism of termination is similar in prokaryotes and eukaryotes, the interactions between the class 1 and class 2 factors have some differences. The class 1 factors RF1 and RF2 recognize the termination codons and activate the ribosome to hydrolyze the peptidyl tRNA.
Cleavage of polypeptide from tRNA takes place by a reaction analogous to the usual peptidyl transfer, except that the acceptor is H O instead of aminoacyl-tRNA. At this point RF1 or RF2 is released from the ribosome by the class 2 factor RF3, which is related to EF-G. RF3-GDP binds to the ribosome before the termination reaction occurs, and the GDP is replaced by GTP. This enables RF3 to contact the ribosomal GTPase center, where it causes RF1 or RF2 to be released when the polypeptide chain is terminated.
RF3 resembles the GTP-binding domains of EF-Tu and EF-G, and RF1 and RF2 resemble the C-terminal domain of EF-G, which mimics tRNA. This suggests that the release factors utilize the same site that is used by the elongation factors. Figure 1 illustrates the basic idea that these factors all have the same general shape and bind to the ribosome successively at the same
site (basically the A site or a region extensively overlapping with it).

FIGURE 1. Molecular mimicry enables the EF-Tu–tRNA complex, the translocation factor EF-G, and the release factors RF1/2-RF3 to bind to the same ribosomal site. RRF is the ribosome recycling factor.
The eukaryotic class 1 release factor, eRF1, is a single protein that recognizes all three termination codons. Its sequence is unrelated to the bacterial factors. It can terminate translation in vitro without the class 2 factor, eRF2, although eRF2 is essential in yeast in vivo. The structure of eRF1 follows a familiar theme; Figure 2 shows that it consists of three domains that mimic the structure of tRNA.

FIGURE 2. The eukaryotic termination factor eRF1 has a structure that mimics tRNA. The motif GGQ at the tip of domain 2 is essential for hydrolyzing the polypeptide chain from tRNA.
An essential motif of three amino acids, GGQ, is exposed at the top of domain 2. Its position in the A site corresponds to the usual location of an amino acid on an aminoacyl-tRNA. This positions it to use the glutamine (Q) to position H2O to substitute for the amino acid of aminoacyl-tRNA in the peptidyl transfer reaction. Figure 3 compares the termination reaction with the usual peptide transfer reaction. Termination transfers a hydroxyl group from H2O, thus effectively hydrolyzing the peptide–tRNA bond.

FIGURE 3. Peptide transfer and termination are similar reactions in which a base in the peptidyl transfer center triggers a transesterification reaction by attacking an N–H or O–H bond, releasing the N or O to attack the link to tRNA.
Mutations in the RF genes reduce the efficiency of termination, as seen by an increased ability to continue translation past the termination codon. Overexpression of RF1 or RF2 increases the
efficiency of termination at the codons on which it acts. This suggests that codon recognition by RF1 or RF2 competes with aminoacyl-tRNAs that erroneously pair with the termination codons. The release factors recognize their target sequences very efficiently.
The termination reaction releases the completed polypeptide but leaves a deacylated tRNA and the mRNA still associated with the ribosome. Figure 4 shows that the dissociation of the remaining components (tRNA, mRNA, 30S, and 50S subunits) requires the ribosome recycling factor (RRF). RRF acts together with EF-G in a reaction that uses hydrolysis of GTP. As for the other factors involved in release, RRF has a structure that mimics tRNA, except that it lacks an equivalent for the 3′ amino acid–binding region. IF-3 is also required. RRF acts on the 50S subunit and IF-3 acts to remove deacylated tRNA from the 30S subunit. Once the subunits have separated, IF-3 remains necessary, of course, to prevent their reassociation.

FIGURE 4. The RF (release factor) terminates translation by releasing the polypeptide chain. The RRF (ribosome recycling factor) releases the last tRNA, and EF-G releases RRF, causing the ribosome to dissociate.
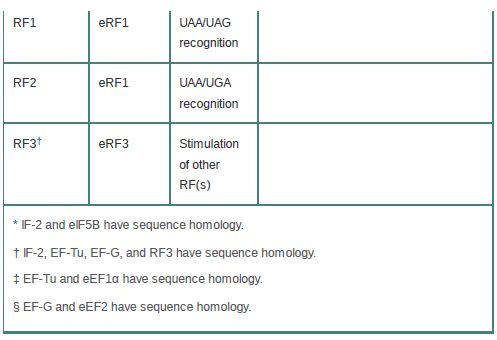
Table 1 compares the functional and sequence homologies of the prokaryotic and eukaryotic translation factors.
TABLE .1 Functional homologies of prokaryotic and eukaryotic translation factors.




|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|