


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Splicing Is Temporally and Functionally Coupled with Multiple Steps in Gene Expression |
|
|
|
Read More
Date: 2-6-2021
Date: 3-1-2016
Date: 1-3-2021
|
Splicing Is Temporally and Functionally Coupled with Multiple Steps in Gene Expression
KEY CONCEPTS
- Splicing can occur during or after transcription.
- The transcription and splicing machineries are physically and functionally integrated.
Splicing is connected to mRNA export and stability control.
- Splicing in the nucleus can influence mRNA translation in the cytoplasm.
Pre-mRNA splicing has long been recognized to take place cotranscriptionally, though the two reactions can take place separately in vitro and have been studied as separate processes in gene expression. Major experimental evidence supporting cotranscriptional splicing came from the observations that many splicing events are completed before the completion of transcription. In general, introns near the 5′ end of the gene are removed during transcription, but introns near the end of the gene can be processed either during or after transcription.
Besides temporal coupling between transcription and splicing, there are probably other reasons for these two key processes to be linked in a functional way. Indeed, the machineries for 5′ capping, intron removal, and even polyadenylation at the 3′ end show physical interactions with the core machinery for transcription. A common mechanism is to use the large C-terminal domain of the largest subunit of Pol II (known as CTD) as a loading pad for various RNA-processing factors, although in most cases it is yet to be defined whether the tethering is direct or mediated by some common protein or even RNA factors .
Such physical integration would ensure efficient recognition of emerging splicing signals to pair adjacent functional splice sites during transcription, thus maintaining a rough order of splicing from the 5′ to 3′ direction. The recognition of the emerging splicing signals by the RNA-processing factors and enzymes associated with the elongation Pol II complex would also allow these factors to compete effectively with other nonspecific RNA-binding proteins, such as hnRNP proteins, that are abundantly present in the nucleus for RNA packaging. If RNA splicing benefits from transcription, why not the other way around? In fact, increasing evidence has suggested so; as illustrated in FIGURE 1, the 5′ capping enzymes seem to help overcome initial transcriptional pausing near the promoter; splicing factors appear to play some roles in facilitating transcriptional elongation; and the 3′ end formation of mRNA is clearly instrumental to transcriptional termination . Thus, transcription and RNA processing are highly coordinated in multicellular eukaryotic cells.
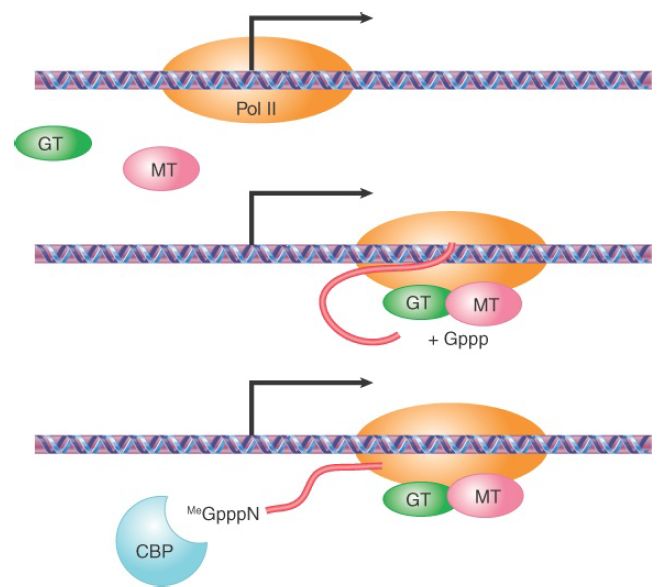
FIGURE 1. Coupling transcription with the 5′ capping reaction. Pol II transcription is initially paused near the transcription start point. Both guanylyl-transferase (GT) and 7-methyltransferase (MT) are recruited to the Pol II complex to catalyze 5′ capping, and the cap is bound by the cap-binding protein complex at the 5′ end of the nascent transcript. These reactions allow the paused Pol II to enter the mode of productive elongation.
RNA processing is functionally linked not only to the upstream transcriptional events but also to downstream steps, such as mRNA export and stability control. It has been known for a long time that intermediately processed RNA that still contains some introns cannot be exported efficiently, which may be due to the retention effect of the spliceosome in the nucleus. Splicingfacilitated mRNA export can be demonstrated by nuclear injection of intronless RNA derived from cDNA or pre-mRNA that will give rise to identical RNA upon splicing. The RNA that has gone through the splicing process is exported more efficiently than the RNAderived from the cDNA, indicating that the splicing process helps mRNA export.
As illustrated in FIGURE 2, a specific complex, called the exon junction complex (EJC), is deposited onto the exon–exon junction. This complex appears to directly recruit a number of RNA-binding proteins implicated in mRNA export. Apparently, these mechanisms may act in synergy to promote the export of mRNA coming out of transcription and the cotranscriptional RNA-splicing apparatus. This process may start early in transcription. The cap binding CBP20/80 complex appears to directly bind to the mRNA export machinery (the TREX complex) in a manner that depends on splicing to remove the first intron near the 5′ end to facilitate mRNA export. A key factor in mediating mRNA export is REE (also named Aly, Yra1
in yeast), which is part of the EJC and can directly interact with the mRNA transporter TAP (Mex67 in yeast), as shown in FIGURE 3.

FIGURE 2 The exon junction complex (EJC) is deposited near the splice junction as a consequence of the splicing reaction.
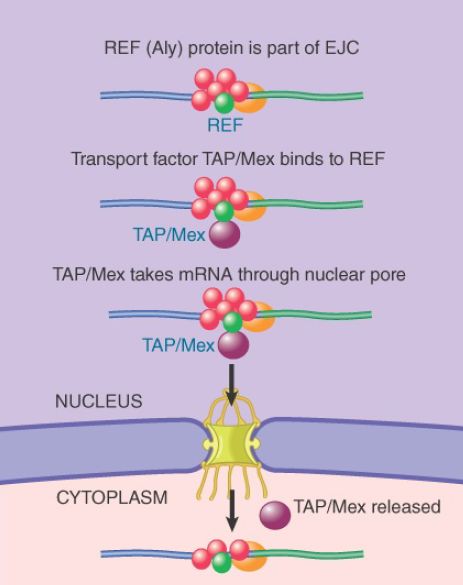
FIGURE 3 An REF protein (shown in green) binds to a splicing factor and remains with the spliced RNA product. REF binds to a transport protein (shown in purple) that binds to the nuclear pore.
The EJC complex has an additional role in escorting mRNA out of the nucleus, which has a profound effect on mRNA stability in the cytoplasm. This is because an EJC that has retained some aberrant mRNAs can recruit other factors that promote decapping enzymes to remove the protective cap at the 5′ end of the mRNA.
As illustrated in FIGURE 4, the EJC is normally removed by the scanning ribosome during the first round of translation in the cytoplasm. If, however, for some reason a premature stop codon is introduced into a processed mRNA as a result of point mutation or alternative splicing (see the next section, titled Alternative Splicing Is a Rule, Rather Than an Exception, in Multicellular Eukaryotes), the ribosome will fall off before reaching the natural stop codon, which is typically located in the last exon. The inability of the ribosome to strip off the EJC complex deposited after the premature stop codon will allow the recruitment of decapping enzymes to induce rapid degradation of the mRNA. This process is called nonsense-mediated mRNA decay (NMD), which represents an mRNA surveillance mechanism that prevents translation of truncated proteins from the mRNA that carries a premature stop codon .

FIGURE 4. The EJC complex couples splicing with NMD. The EJC can also recruit Upr proteins if it remains on the exported mRNA. After nuclear export, EJC should be tripped off by the scanning ribosome in the first round of translation. If an EJC remains on the mRNA because of a premature stop codon in the front, which releases the ribosome, the EJC will recruit additional proteins, such as Upf, which will then recruit the decapping enzyme (DCP). This will induce decapping at the 5′ end and mRNA degradation from the 5′ to 3′ direction in the cytoplasm.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|