


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 31-3-2021
Date: 10-12-2015
Date: 2-5-2021
|
The Bacterial Genome Is (Usually) a Single Circular Replicon
KEY CONCEPTS
- Bacterial replicons are usually circles that replicate bidirectionally from a single origin.
- The origin of Escherichia coli, oriC, is 245 base pairs (bp) in length.
Prokaryotic replicons are usually circular, so that the DNA forms a closed circle with no free ends. Circular structures include the bacterial chromosome itself, all plasmids, and many bacteriophages, and are also common in chloroplasts and mitochondrial DNAs. FIGURE 1 summarizes the stages of replicating a circular chromosome. After replication has initiated at the origin, two replication forks proceed in opposite directions. The circular chromosome is sometimes described as a θ structure at this stage, because of its appearance. An important consequence of circularity is that the completion of the process can generate two chromosomes that are linked because one passes through the other (they are said to be catenated), and specific enzyme systems may be required to separate them .
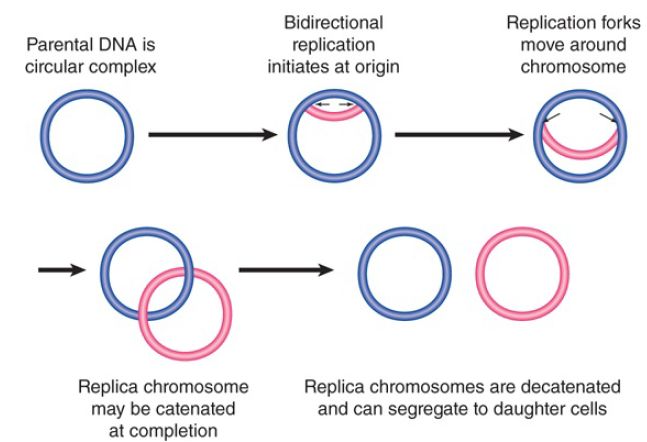
FIGURE 1. Bidirectional replication of a circular bacterial chromosome is initiated at a single origin. The replication forks move around the chromosome. If the replicated chromosomes are catenated, they must be disentangled before they can segregate to daughter cells.
The genome of E. coli is replicated bidirectionally from a single unique site called the origin, identified as the genetic locus oriC. Two replication forks initiate at oriC and move around the genome at approximately the same speed to a special termination region . One interesting question is this: What ensures that the DNA is replicated right across the region where the two forks meet?
What happens when a replication fork encounters a protein bound to DNA? We assume that repressors, for example, are displaced and then rebind. A particularly interesting question is what happens when a replication fork encounters an RNA polymerase engaged in transcription. A replication fork moves 10 times faster than RNA polymerase. Under the best of conditions, in log phase growth, collisions between the replication machinery and RNA polymerase do occur. In times of stress, such as amino acid starvation, it increases. A set of transcription factors acting as elongation factors interact with RNA polymerase to facilitate replication read through by removing transcription roadblocks, but this requires active transcription. It is not yet clear what the mechanism of action is. Most active transcription units are oriented so that they are expressed in the same direction as the replication fork that passes them. Many exceptions comprise small transcription units that are infrequently expressed. The difficulty of generating inversions containing highly expressed genes suggests that head-on encounters between a replication fork and a series of transcribing RNA polymerases might be lethal.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|