


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 15-11-2020
Date: 30-12-2015
Date: 29-11-2015
|
Antibody Phage Display
Phage display was the first antibody display technology to be developed and is the one most widely used today . Antibody phage display resulted from concurrent progress in prokaryotic expression of antibody fragments, PCR cloning of antibody gene repertoires and display of peptides and proteins on filamentous bacteriophages. By applying PCR and V region-specific primers to RNA prepared from human peripheral blood lymphocytes or mouse splenocytes, it is possible to amplify repertoires of VH and VL genes . The V gene repertoires can then be either separately cloned sequentially or spliced by overlap extension to create human or murine scFv or Fab gene repertoires . An alternative to the use of naturally occurring V gene repertoires is to synthesize V genes in vitro using cloned or synthetic V gene segments and random oligonucleotides encoding part of the antigen combining site.72,74 Using this approach, it is possible to ensure that the potential V gene repertoire diversity is large enough that a library will only contain a single member of each sequence. In contrast, there may be considerable duplication of sequences using naturally occurring V genes.
The filamentous bacteriophages that infect E. coli, such as M13 and fd phage, are the phages used for antibody display. Such phage are singlestranded DNA viruses where the phage contains one copy of the viral DNA which is covered by a protein coat. The coat consists of approximately 3000 copies of the major coat protein pVIII, three to five copies of the minor coat protein pIII and the coat proteins pVII and pIX (Figure 2). scFv and Fab antibody fragments have been successfully displayed on pIII, pVII, pVIII and pIX75–77 (Figure 2). Antibody phage display is accomplished by cloning antibody genes into a vector where they are in-frame with one of the coat proteins. They are then
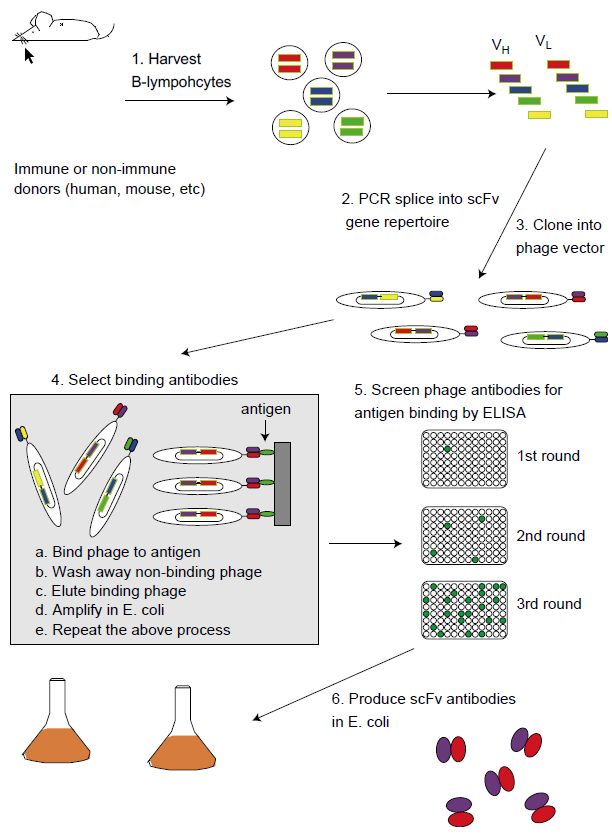
Figure 1 For phage display, B-cells are isolated from immunized mice (as in panel A) or naive or immunized humans. Heavy and light chain V genes (shaded bars) are amplified by PCR and assembled as single-chain Fv antibody genes (scFvs). Alternatively, rearranged V genes can be generated entirely in vitro from cloned V segments and synthetic oligonucleotides. The repertoire of scFv genes are cloned into a phage display vector, where the encoded scFv proteins (colored ovals) are displayed as fusion proteins to one of the phage coat proteins. The phage contain the appropriate scFv gene within. Multiple rounds of selection with immobilized antigen allow the isolation of even rare antigen binding phage antibodies, which are identified by ELISA. Native scFvs can be expressed from E. coli and purified for characterization and use in assays.
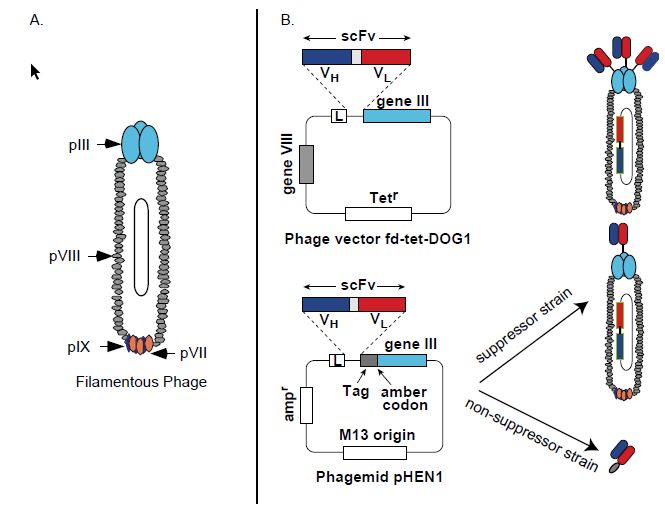
Figure 2 Features of phage display vectors. (A) Illustration of phage proteins pIII, pVII, pVIII and pIX, which can be used for antibody fragment display. (B) Representative phage and phagemid vectors. Phage vector fd-tet-DOG1 (top panel) and phagemid vector pHEN1 (bottom panel). Both vectors display scFv (VH–linker–VL) as fusions to the amino terminus of the pIII protein. Both vectors have leader sequences (pelB or gene III) to direct the expressed fusion protein to the bacterial periplasm. In phage fdtet- DOG1, all copies of pIII are scFv fusions, leading to 3–5 copies displayed per phage particle. With phagemid pHEN1, expression in suppressor strains of E. coli allows the amber codon following the scFv-tag to be read as a glutamine, causing the scFv to be fused to the pIII protein. In phagemids, both wild-type pIII (from the helper phage) and fusion pIII (from the phagemid) compete for inclusion in the viral particle. In nonsuppressers E. coli strains, the scFv is expressed as a soluble protein with a myc epitope tag for detection of binding in ELISA.
expressed as fusion proteins on the phage surface (Figure 2). The phage provides a physical link between the antibody fragment on its surface and the gene that encodes the antibody contained within the phage. The first example of antibody phage display was reported in 1990 by McCafferty, who demonstrated that an scFv fragment of the antilysozyme Mab D1.3 could be functionally displayed on pIII in the phage vector fd and that the scFv retained antigen binding (Figure 2).
Moreover, phage displaying the scFv could be enriched from a mixture with wild-type phage by affinity chromatography on a lysozyme column. Enrichment factors of 103 for one round of selection and 106 for two rounds of selection were achieved. A large number of vector systems have subsequently been described for the display of antibody fragments (Figure 2). The vectors differ primarily with respect to the type of antibody fragment displayed, Fabor scFv. the fusion partner pIII or pVIII and whether the vector is a phage or phagemid. The most widely used vectors by far are those that display scFv or Fab antibody fragments as pIII fusions in phagemid vectors (Figure 2). pIII display appears to be more robust than display on the other phage proteins; phagemids result in much higher transformation efficiencies, leading to larger antibody library sizes. Use of a phagemid vector results in ‘monovalent’ antibody fragment display compared with display in true phage vectors such as fd (Figure 9.6). This is because phagemid vectors do not contain the genes for phage proteins other than pIII. The other genes, and regulatory DNA, are provided in trans by infecting E. coli with helper phage. Wild-type pIII from the helper phage competes with the antibody fragment–pIII fusion for incorporation into the phage coat.
scFv or Fab gene repertoires can be cloned into phage or phagemid display vectors to create what are called ‘libraries’ of phage, each with a different antibody fragment on its surface and the gene for the antibody fragment inside ( Figure 1). Given the high transformation of E. coli, it is possible to create libraries containing millions to billions of different antibodies (Figure 1). Rare antibodies binding specific antigens can be isolated from non-binding antibodies by a range of different types of affinity chromatography. For example, antigen can be adsorbed on a plastic plate and then phage incubated with immobilized antigen, non-binding phage removed by washing and bound phage eluted. A single round of selection will result in a 20–1000-fold enrichment for binding phage. Eluted phage are used to infect E. coli, which produce more phage for the next round of selection. Repetition of the selection process makes it possible to isolate binding phage present at frequencies of less than one in a billion.
Adsorption of protein antigens on plastic may lead to partial or complete antigen denaturation. The problem of antigen denaturation can be overcome by selecting a soluble antigen in solution. The antigen can be chemically tagged, for example by biotinylation, and captured along with bound phage using avidin or streptavidin magnetic beads.
Alternatively, the antigen can be genetically tagged with a hexahistidine tag with capture on Ni-NTA agarose or expressed as GST or maltose binding protein fusions with capture on either glutathione or maltose columns. Selections can be performed on more complex mixtures of antigen, including intact cells, provided that measures are taken to prevent enrichment of phage antibodies which bind to non-relevant cell surface antigens. Cell surface antigen specific antibodies have been isolated by selecting phage antibody libraries on adult erythrocytes,81 fetal erythrocytes, lymphocytes, melanoma cells and breast tumor cells.85 It has also proven possible to select directly phage antibodies that trigger receptor-mediated endocytosis. Such antibodies can be used for the targeted delivery of therapeutics to the cytosol.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|