

علم الكيمياء

تاريخ الكيمياء والعلماء المشاهير

التحاضير والتجارب الكيميائية

المخاطر والوقاية في الكيمياء

اخرى

مقالات متنوعة في علم الكيمياء

كيمياء عامة


الكيمياء التحليلية

مواضيع عامة في الكيمياء التحليلية

التحليل النوعي والكمي

التحليل الآلي (الطيفي)

طرق الفصل والتنقية


الكيمياء الحياتية

مواضيع عامة في الكيمياء الحياتية

الكاربوهيدرات

الاحماض الامينية والبروتينات

الانزيمات

الدهون

الاحماض النووية

الفيتامينات والمرافقات الانزيمية

الهرمونات


الكيمياء العضوية

مواضيع عامة في الكيمياء العضوية

الهايدروكاربونات

المركبات الوسطية وميكانيكيات التفاعلات العضوية

التشخيص العضوي

تجارب وتفاعلات في الكيمياء العضوية


الكيمياء الفيزيائية

مواضيع عامة في الكيمياء الفيزيائية

الكيمياء الحرارية

حركية التفاعلات الكيميائية

الكيمياء الكهربائية


الكيمياء اللاعضوية

مواضيع عامة في الكيمياء اللاعضوية

الجدول الدوري وخواص العناصر

نظريات التآصر الكيميائي

كيمياء العناصر الانتقالية ومركباتها المعقدة


مواضيع اخرى في الكيمياء

كيمياء النانو

الكيمياء السريرية

الكيمياء الطبية والدوائية

كيمياء الاغذية والنواتج الطبيعية

الكيمياء الجنائية


الكيمياء الصناعية

البترو كيمياويات

الكيمياء الخضراء

كيمياء البيئة

كيمياء البوليمرات

مواضيع عامة في الكيمياء الصناعية

الكيمياء الاشعاعية والنووية
Proteins that sense Cu and Zn levels
المؤلف:
Peter Atkins, Tina Overton, Jonathan Rourke, Mark Weller, and Fraser Armstrong
المصدر:
Shriver and Atkins Inorganic Chemistry ,5th E
الجزء والصفحة:
771
2025-10-28
706
Proteins that sense Cu and Zn levels
Key point: Cu and Zn are sensed by proteins with binding sites specially tailored to meet the specific coordination preferences of each metal atom. The levels of Cu in cells are so strictly controlled that almost no uncomplexed Cu is present. An imbalance in Cu levels is associated with serious health problems such as Menkes disease (Cu deficiency) and Wilson’s disease (Cu accumulation). Most of what we know about how Cu levels are sensed and converted to cell signals stems from studies on the E. coli system, which involves a transcription factor called CueR (Fig. 27.53). This protein binds Cu(I) with high selectivity, although it also binds Ag(I) and Au(I). Metal coordination causes a conformational change that enables CueR to bind to DNA at a receptor site that controls transcription of an enzyme known as CopA, which is an ATP-driven Cu pump. CopA is located in the cytoplasmic membrane and exports Cu into the periplasm. In CueR, the Cu(I) is coordinated by two cysteine-S atoms arranged in a linear coordination geometry. Titrations using CN as a buffer show that Cu is bound with a dissociation constant of approximately 10 21. As may be understood by reference to Section 7.3, this ligand environment leads to remarkably selective binding for d10 ions, and measurements with Ag and Au show that these ions are taken up with similar affinities. Most of our current insight about Zn sensing, as for Cu, is provided by studies of bacterial systems. The major difference with respect to Cu is that although Zn is also coordinated (mainly) by cysteine thiolates, the geometry is tetrahedral rather than linear. E. coli contains a Zn2-sensing transcription factor known as ZntR that is closely related to CueR. The factor ZntR contains two Zn-binding domains each of which coordinates a pair of Zns using cysteine and histidine ligands. The surrounding protein fold is shown in Fig. 27.53, for comparison with CueR. The extent to which these dynamic Zn-binding sites can be identified with zinc fingers remains unclear.
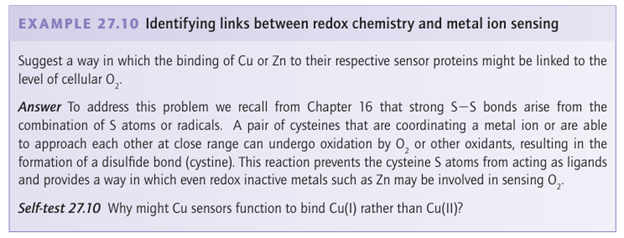
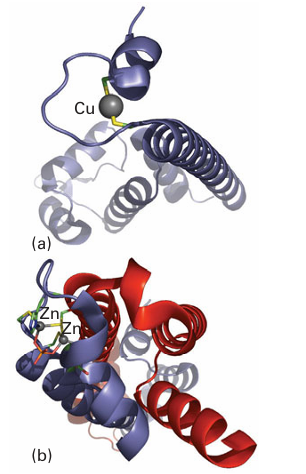
Figure 27.53 Comparison of (a) the Cu- and (b) the Zn-binding sites in the respective transcription factors CueR and ZntR. Note how Cu(I) is recognized by a linear binding site (to two cysteines) whereas Zn is recognized by an arrangement of Cys and His ligands that binds two Zn (II) atoms together with a bridging phosphate group.
 الاكثر قراءة في مواضيع عامة في الكيمياء العضوية
الاكثر قراءة في مواضيع عامة في الكيمياء العضوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















