x

هدف البحث
بحث في العناوين
بحث في اسماء الكتب
بحث في اسماء المؤلفين

اختر القسم
موافق


النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الحيوية

مواضيع عامة في المضادات الحيوية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
MicroRNAs Are Widespread Regulators in Eukaryotes
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
17-6-2021
2234
MicroRNAs Are Widespread Regulators in Eukaryotes
KEY CONCEPTS
- Eukaryotic genomes encode many short RNA molecules called microRNAs (miRNAs).
- Piwi-interacting RNAs (piRNAs) regulate gene expression in germ cells and act to silence transposable elements.
- Small interfering RNAs (siRNAs), or silencing RNAs, are complementary to viruses and transposable elements.
Eukaryotes, like bacteria, use RNAs to regulate gene expression. Noncoding RNAs are used to control gene expression in the nucleus at the level of DNA; in many cases the expression and function of these RNAs are inextricably linked to chromatin structure. Transcription of tandemly repeated, simple sequence satellite heterochromatic DNA is required for the formation of heterochromatin itself . This section focuses mainly on control in the cytoplasm at the level of the mRNA. As will be described, the eukaryotic mechanisms, though related to the bacterial mechanisms, are very different. Like bacteria, eukaryotes use RNA to regulate gene expression. Note, though, that attenuation is not possible in eukaryotes (as it is in E. coli), because the nuclear membrane separates the processes of transcription and translation. Given that eukaryotic mRNA is so much more stable than bacterial mRNA, with an average half-life of hours as opposed to minutes, much more translation-level control is used in eukaryotes, both at the level of translation initiation and mRNA stability control itself in the cytoplasm (see the chapter titled mRNA Stability and Localization).
Numerous classes of small noncoding RNAs have been identified in eukaryotes, besides the major 5S rRNA and tRNAs. Some of these have been described elsewhere, such as the different classes of guide RNAs that are involved in RNA splicing, editing, and modification (see the chapters titled RNA Splicing and Processing and Catalytic RNA).
Very small RNAs—microRNAs, or miRNAs—are gene-expression regulators found in most, if not all, eukaryotes. These bear some resemblance to their bacterial sRNA counterparts, but they are typically smaller and their mechanism of action is different. The human genome has an estimated 1,500 genes that encode miRNAs that participate in RNA interference (RNAi), about half from the introns of coding genes, and about half from large ncRNAs. Even more interesting, miRNAs can originate from pseudogenes—supposedly inactive genelike regions that were thought to have no function. This is a general mechanism to repress gene expression, usually (but not always) at the level of translation. These miRNAs go by a number of names and are sometimes called short temporal RNA, or stRNA, because many are involved in development. Some miRNAs have also been shown to affect transcription initiation by binding to the gene’s promoter. It is estimated that these miRNAs control thousands of mRNAs, perhaps as much as 90% of the gene total, at all stages of development. Each miRNA may have
hundreds of target mRNAs. A given mRNA may be the target of multiple miRNAs.
Piwi-interacting RNAs, piRNAs, are a special class of miRNA found in germ cells. Another type of very small RNA is siRNA (small interfering RNA), which is typically produced during a virus infection. Both piRNAs and siRNAs can be used to control the expression of transposable elements. In fact, this may be how these small RNAs originated and evolved. These RNAs have multiple origins and multiple mechanisms of synthesis and processing. Most are produced as larger precursor RNAs that are processed and cleaved to the correct size and then delivered to their target.
The miRNAs used in RNAi are produced as large RNA primary transcripts called pri-miRNAs that are self-complementary and can automatically fold into a double-strand hairpin structure, usually with some imperfect base pairing. The pri-miRNA is processed in a twostep reaction (shown in FIGURE 1). The first step is catalyzed by Drosha, an RNase III superfamily member endonuclease, in the nucleus. Drosha reduces the pri-RNA to about a 70-bp, hairpinshaped precursor fragment, pre-miRNA, which has a phosphate group at the 5′ end. This cleavage determines the 5′ and 3′ ends of the precursor.
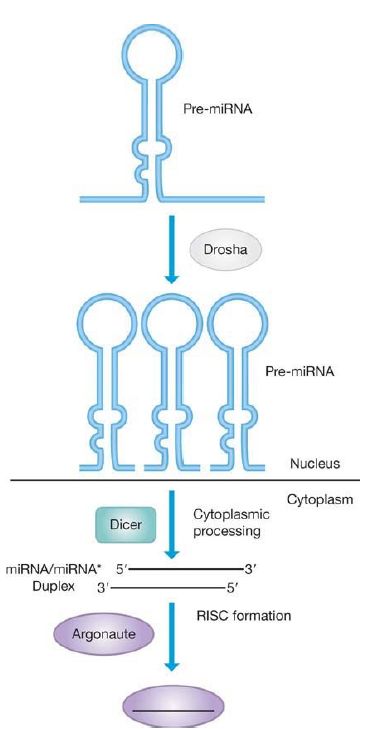
FIGURE 1. miRNAs are generated by processing from a precursor pre-miRNA by the enzyme Drosha. The pre-miRNA is then processed by the enzyme Dicer for assembly into the Argonaute complex.
Data from I. Slezak-Prochazka, et al. RNA 16 (2010): 1087–1095 and S. Bajan and G. Hutvagner, Mol. Cell 44 (2011): 345–347.
After export from the nucleus to the cytoplasm, the second step, pre-miRNA to miRNA, is catalyzed by a second RNase III family member, Dicer, by counting from the 3′ end to produce a short, double-stranded segment that is approximately 22 bp. The miRNA now has a short, two-nucleotide single-stranded 3′ end, which is then usually modified by adding a 2′-O-methyl group for stability. Dicer has an N-terminal helicase activity, which enables it to unwind the double-stranded region, and two nuclease domains that are also related to the bacterial RNase III. Related enzymes are nearly universal in eukaryotes. In plants, the Dicer-like enzyme performs both the pri-miRNA and pre-miRNA processing steps in the nucleus.
Extensive modifications, beyond the standard 2′-O-methylation, are possible. Some pri-miRNAs can undergo RNA editing by the enzyme ADAR, which converts adenosine to inosine. This can result in altered target specificity. miRNAs can also undergo uridylation or adenylation at the 3′ end. Short oligo-U tracts are a signal for degradation, whereas oligo-A tracts (and 2-Omethylation) have the opposite effect.
These short, double-stranded RNA fragments are delivered to, or loaded onto, a complex called RISC (RNA-induced silencing complex). Proteins in the Argonaute (Ago) family are components of this complex and are required for the final processing to a single strand, by the elimination of the passenger strand, which is denoted as miRNA*. RISC then (usually) delivers the miRNA to the 3′ untranslated region (UTR) of its target mRNA. Humans have 8 Ago family members, Drosophila has 5, plants have 10, and Caenorhabditis elegans has 26. These proteins have an ancient origin and are found in bacteria, archaea, and eukaryotes (this system is absent in the yeast Saccharomyces cerevisiae but is present in some of its close relatives). The degree of base pairing and the sequence of the ends (determined by Dicer cleavage) of the duplex dictate which of the multiple Ago family members picks up the RNA duplex and which strand is selected as the passenger strand to be degraded, as shown in FIGURE 2 . The RISC complex is now in a position to use the mature miRNA to guide it to its target mRNA. Selection of the class of target by RISC lies with the specific Argonaute protein; the specific RNA target itself is determined by the miRNA.
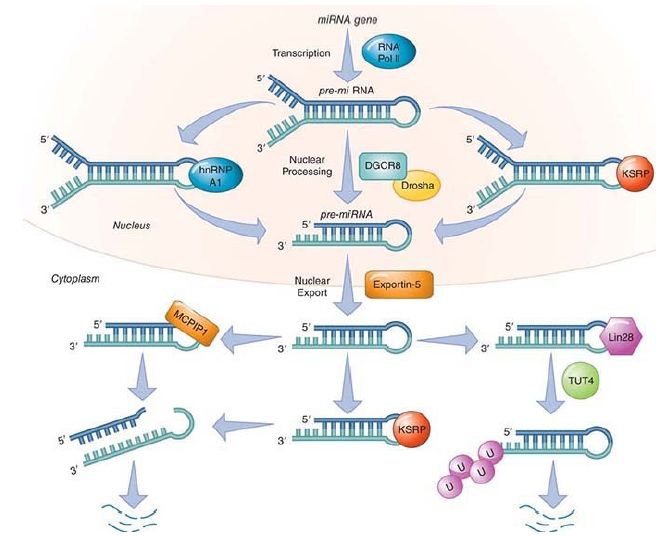
FIGURE 2. Processing and regulation of miRNA processing via the loop. The basic steps of the canonical miRNA processing from transcription are shown. Several proteins that regulate this process by directly binding to the loop sequence of miRNA(s) are indicated. The function of MCPIP1 and Lin28 are negative regulators of a set of miRNAs. MCPIP1 cleaves the loop, which leads to degradation of the set that it regulates. Lin28 recruits a uridylyl transferase enzyme, which adds a poly(U) tail leading to degradation. KSRP, another regulatory factor, is a positive regulator.
Data from S. Bajan and G. Hutvagner, Mol. Cell 44 (2011): 345–347.
A germline subset of miRNA is the Piwi-interacting RNA, (originally P element–induced wimpy testis). In Drosophila, these are sometimes called rasiRNAs (repeat-associated siRNAs). These are so named because they interact with a different subfamily member of the Ago class proteins, known as Piwi (also called Miwi in mice and Hiwi in humans). Piwi-class proteins are only found in metazoan organisms (multicellular eukaryotes). In addition, the piRNAs are somewhat longer than miRNAs, ranging from 24 to 31 nucleotides, and also 2′-O-methylated at their 3′ end. piRNAs are found in giant tandem clusters, sometimes with tens of thousands of copies. The processing pathway has not yet been determined, but it is probably similar to that of the miRNAs. They are delivered to different Ago family members than miRNAs, including the Piwi, Aubergine, and Ago3 proteins.
The function of the piRNAs is also different from miRNAs. Their primary function is nuclear, repressing the expression of transposable elements, preserving genome integrity, and controlling chromatin structure . The mechanism whereby piRNAs affect chromatin control is reminiscent to what was described in the chapter Epigenetics II. In the mouse (and in mammals in general) certain genes show parental origin-specific expression due to DNA methylation patterns in differentially methylated regions (DMRs). Methylation of the gene Rasgrf1 is controlled through its DMR, which contains both long interspersed elements (LINEs) and short
interspersed elements (SINEs). These are transcribed into piRNAs and long ncRNAs that then serve as a scaffold for the enzymes that methylate and repress transcription from Rasgrf1.
Only a few of the piRNAs are complementary to transposable elements. Most map to single-copy DNA, both genes, and intergenic regions. In Drosophila, it is maternally inherited piRNAs that provide protection against transposon activation to the female from P element–mediated hybrid dysgenesis .
siRNAs have a different origin. These are derived from viral infections, which typically transcribe both genomic strands to produce complementary double-stranded RNAs. These large, double-stranded RNAs are processed by Dicer in a manner similar to that of the miRNAs described earlier and are delivered to RISC. siRNAs use a different Ago family member (and therefore a different RISC). siRNAs are also derived from transcription of transposable elements and are used to silence them. An interesting feature of siRNAs is that they have the ability to spread from cell to cell throughout an organism, a useful feature to have during a viral infection. This phenomenon is very common in plants and has also been seen in C. elegans. This process can be amplified in these organisms by an RNA-dependent RNA polymerase. Humans and Drosophila may not possess this polymerase enzyme.