DNA Chips
The start of the Human Genome Project in the late 1980s refocused scientists’ attention on new, high-throughput technologies for handling and analyzing DNA. Several technologies developed by physicists were evaluated for their potential use in the study of biomolecules. For example, mass spectrometry techniques and the scanning tunneling microscope are now used for analyzing and sequencing DNA. Another technology developed by physicists and applied to biology is the microchip. The semiconductor industry manufactured silicon chips with smaller and smaller features, which allows for greater numbers of operations with a chip of a standardized size. This benefit of miniaturization was applied to the biological sciences in the form of multiparallel arrays of DNA fragments on a small chip. Arraying of DNA or RNA samples onto nitrocellulose or nylon membranes for hybridization studies is a very common analytical procedure in a molecular biology lab (1). DNA chips or microarrays are, in principle, miniaturized versions of these standard dot-blot techniques. With great precision, thousands of DNA fragments, often amplified by PCR, or DNA oligonucleotides are spotted/printed onto glass microscope slides. In the case of oligonucleotides, they can also be synthesized directly on coated silicon chips. These DNA chips are then used to probe fluorescent-labeled DNA or RNA samples for the presence of specific complementary sequences. DNA chip technology is currently being applied in the areas of monitoring gene expression, polymorphism analysis, gene mutation analysis, and DNA sequencing.
1. Putting Genes on a Chip
The fabrication of high-density arrays on chips for parallel analysis of up to several thousand cDNA clones or PCR fragments on a surface area of approximately 1 cm2 can be achieved by two techniques: microprinting or piezojet dispensing. The high-density arrays are deposited on the flat solid support, usually a glass slide. The key function of the printing or dispensing process is the extraction of a small volume of sample from a reservoir (eg, a microtiter plate) and the deposition of the same amount of sample fluid in each spot, at equivalent locations on each chip. The center-to-center separation of individual spots in high-density arrays is typically 150 to 500 µm. The deposited sample volume ranges form 50 to 400 pl per spot.
Microprinting devices use an array of pins with either hollow steel capillaries or flat tips. The amount of sample deposited at each spot is defined by the capillary forces acting on the droplet, which adheres to the pin after immersion into the sample reservoir. The droplets at the tips are then transferred to the spots in the array by moving the pins close to the chip surface. After each sample, the tip is cleaned by rinsing. The amount of liquid deposited depends on the size of the tip.
The piezojet is a contactless dispensing technique for subnanoliter droplets. It is based on a cylindrical piezo ceramic that compresses a glass capillary with an inner diameter of ~1 mm on application of an electric pulse. A small droplet is then expelled through the funnel-shaped orifice of the capillary. The outlet diameter of the orifice is ~50 µm. Very uniform droplets can be prepared by this method (see Fig. 1).
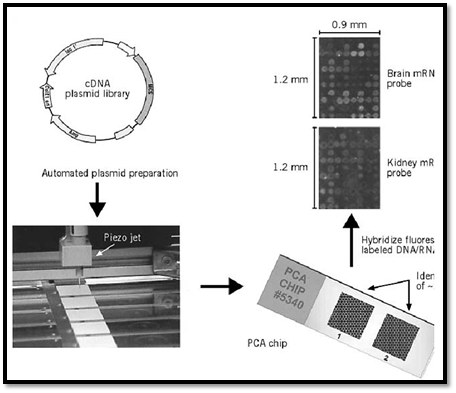
Figure 1. The principle of preparing and applying plasmid chip arrays (PCA), as developed by U. Certa and R. Hochstra Hoffmann–La Roche Ltd, Basel, Switzerland).
The piezojet-dispensing process consists of three steps: (1) Aspiration of an aliquot of sample through the capillary, (2) dispensing of one subnanoliter droplet of sample to equivalent spots on each chip, and (3) discharge of remaining sample volume and cleaning of the piezojet. In order to use the sample material effectively, the aspiration volume of the piezojet should be in the microliter range.
The deposited DNA has to be immobilized on the surface in order to enable the subsequent processing steps. The most commonly used attachment method is to coat the glass slides with polylysine, which holds the negatively charged DNA backbone by electrostatic interactions. Alternative methods, like streptavidin-coated slides to bind biotin-labeled DNA, and various chemical approaches to bind DNA covalently to pretreated glass, are under development.
The most elegant way to attach DNA fragments covalently onto the glass chips has been developed by Fodor and colleagues at Affymetrix (2-4). This method allows light-directed synthesis of thousands of DNA fragments (oligonucleotides) in precise locations on the microchip. To begin the process, linkers modified with a photochemically removable protecting group are attached to the solid substrate. Light is directed through a photolithographic mask, illuminating specific grid squares on the chip and causing photodeprotection, or the removal of the blocking group, in those squares. The chip is then incubated with a nucleotide harboring a photolabile protecting group at the 5′ end. The cycle continues: the chip is exposed to light through the next mask, which activates new grid sites for reaction with the subsequent photoprotected nucleotide. Using the proper set of masks and chemical steps, it is possible to construct a defined collection of oligonucleotides, generally 20 to 25 bases long, each in a predefined position on the array. A standard 1.28-cm2 chip can currently be packaged with about 400,000 individual, well-defined oligonucleotides representing many thousand genes.
2. Monitoring Gene Expression
A bacterial genome, such as that of Escherichia coli, encodes 4288 different genes, the yeast Saccharomyces cerevisiae genome about 6000 genes; Caenorhabditis elegans has about two times more genes, and the human genome may contain approximately 100,000 unique genes. At least partial sequence information for all these genes will be available soon. Sequence information alone, however, is insufficient for a full understanding of gene function and the control of gene expression. Only a subset of all encoded genes is expressed in any given cell; in higher eukaryotes, this subset is smaller than in bacteria or yeast cells. The levels and the timing of gene expression determine the fate of the cells, their reproduction, differentiation, function, communication, and physiology. Methods such as Northern blots, nuclease protection, and RT (reverse transcriptase)- PCR are frequently used to measure gene transcripts, but they have the inherent disadvantage of being serial, analyzing a single messenger RNA at a time. Most comparative techniques, like subtractive hybridization, differential display (5) of amplified mRNA's on gels, and the SAGE (serial analysis of gene expression) method of Velculescu et al. (6), are laborious, often nonreproducible, and not particularly sensitive. Arrays of several thousand genes, the DNA chips, for the first time make it possible to obtain gene expression information on complete genomes (7) quickly, accurately, and efficiently. These gene arrays are in principle reverse Northern blots, where the DNA probes have been immobilized to identify and measure large numbers of mRNA species in parallel. Two prototype DNA microarrays for gene expression monitoring can be described:
2.1. cDNA Microarrays
Brown and his colleagues developed a high-capacity system to monitor the expression of many genes in parallel (8, 9). In short, the complementary DNA of expressed mRNAs from cells is collected, and individual cDNA molecules are isolated and amplified. A microsample of each cDNA from the library is then deposited by high-speed robotic printing on a polylysine-coated glass surface in an array format. Each gene has a unique location in the microarray, which may represent several thousands of genes on a few square centimeter. To compare expression of these genes between two tissues or two cell types, the poly(A)+–mRNA in the two samples is copied into cDNA and labeled with two differently colored fluorescent molecules. Both probe samples are applied to a single microarray and allowed to react with the DNA on the microarray. After the appropriate washing steps, each element of the microarray is scanned for fluorescence intensity from the two colors by a laser fluorescence scanner. The observed fluorescence intensity at each array element is proportional to the number of fluorescent cDNA molecules bound to it, and the ratio of the two colors is an accurate measurement of the relative expression level of the genes in the two tissue or cell samples. Twofold changes in expression can be detected (10). As an illustration, Figure 1 shows a similar DNA microarray, the plasmid chip array (PCA), made by piezojet dispensing in the labs of R. Hochstrasser and U. Certa (F. Hoffmann-La Roche Ltd., Basel, Switzerland).
2.2. Oligonucleotide Microarrays
High-density arrays of oligonucleotide probes represent the other prototype method for mRNA expression monitoring. Sequence information about the expressed genes is used directly to select oligonucleotides and to design the photolithographic masks for combinatorial synthesis of the probes directly on derivatized glass, as described previously. The arrays are intentionally redundant, as they contain collections of pairs of probes for each of the RNAs being monitored. Each probe pair consists of a 20- to 25-mer oligonucleotide that is perfectly complementary to a subsequence of a particular message and a mismatch oligo that is identical except for a single base difference in a central position, which serves as an internal control for hybridization specificity. For array
hybridization experiments, target RNA is prepared from cellular mRNA by incorporating labeled ribonucleotides in an in vitro transcription reaction (11), or by direct chemical attachment (12). After hybridization and washing, fluorescence imaging of the arrays is accomplished with a scanning confocal microscope. Because oligonucleotide probes for each gene are specifically chosen and synthesized in known locations on the arrays, the hybridization patterns and intensities can be interpreted in terms of gene identity and the relative amount of each transcript. This technology has been successfully applied for monitoring gene expression in mammalian cells (11), yeast (7), and bacteria (12), measuring expression levels of less than one copy of mRNA to several hundred copies per cell in one hybridization experiment.
3. Accessing Genetic Diversity
High-density oligonucleotide probe arrays (Fig. 2) can also be applied to a broad range of nucleic acid sequence analysis problems, including pathogen identification, polymorphism detection and sequence checking. The significant instability of internal probe–target mismatches relative to perfect matches (4, 13) is used to design arrays of probes capable of detecting differences between nucleic acid targets. To interrogate the identity of a nucleotide as position X of a known DNA sequence, four oligonucleotides can be offered with the four different choices A, G, T, C in the middle of 15 or so flanking nucleotides that perfectly match the reference sequence. The probe with the highest intensity after hybridization would indicate the identity of the unknown base. This concept can be extended to detect polymorphisms/mutations relative to a characterized consensus sequence. Thus, to screen 1000 nucleotides for polymorphisms/mutations would require 4000 probes. Current applications of this technique include the survey of drug-resistance mutations in HIV-1 reverse transcriptase and proteinase genes, a p53 mutations screen, and a cytochrome P450 allelic variants screen.

Figure 2. The basic principle of oligonucleotide microarrays (courtesy of Affymetrix, Santa Clara, CA).
4. Sequencing by Hybridization
Hybridization can also be used to determine the sequence of unknown DNA, ie, de novo DNA sequencing. Sequencing by hybridization is based on the use of oligonucleotide hybridization to determine the set of constituent subsequences in a DNA fragment. This concept which was spearheaded by Crkvenjakov (14, 15) uses a microarray of all possible n-nucleotide oligomers, eg, 65,635 possible octamers, to identify all the n-mers present in an unknown DNA sequence. Powerful computational approaches are then used to assemble the complete sequence from the measured hybridization patterns. High-density oligonucleotide probe array technology shows significant promise in enabling de novo sequencing in a fast and reliable manner.
References
1. G. G. Lennon and H. Lehrach (1991) Trends Genet. 7, 314–317.
2. S. P. A. Fodor, J. L. Read, M. C. Pirrung, L. Stryer, A. T. Lu, and D. Solas (1991) Science 251, 767-773.
3. S. P. A. Fodor, R. P. Rava, X. C. Huang, A. C. Pease, C. P. Holmes, and C. L. Adams (1993( Nature 364, 555–556.
4. A. C. Pease, D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. A. Fodor (1994( Proc. Natl. Acad. Sci. USA 91, 5022–5026.
5. P. Liang and A. B. Pardee (1992) Science 257, 967–971.
6. V. E. Velculescu, L. Zhang, B. Vogelstein, and K. W. Kinzler (1995) Science 270, 484–487.
7. L. Wodicka, H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart (1997) Nature Biotech. 15, 1359-1367.
8. M. Schena, D. Shalon, R. W. Davis, and P. O. Brown (1995) Science 270, 467–470.
9. D. Shalon, S. J. Smith, and P. O. Brown (1996) Genome Res. 6, 639–645.
10. M. Schena, D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis (1996) Proc. Natl. Acad. Sci. USA 93, 10614–10619.
11. D. J. Lockhart, H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown (1996) Nature Biotech. 14, 1675–1680.
12. A. de Saizieu, U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous (1998) Nature Biotech. 16,45-48.
13. R. J. Lipshutz, D. Morris, M. Chee, E. Hubbell, N. S. Kozal, N. Shen, R. Young, and S. P. A. Fodor (1995) BioTechniques 19, 442–447.
14. R. Drmanac, I. Labat, I. Brukner, and R. Crkvenjakov (1989) Genomics 4, 114–128.
15. R. Drmanac, S. Drmanac, Z. Strezoska, T. Paunesku, I. Labat, M. Zeremski, J. Snoddy, W. K. Funkhouser, B. Koop, L. Hood, and R. Crkvenjakov (1993) Science 260, 1649–1652.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


