

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Presence/absence method MLG/FSIS/USDA 2011 for Salmonella in foods
المؤلف:
SILVA, N.D .; TANIWAKI, M.H. ; JUNQUEIRA, V.C.A.; SILVEIRA, N.F.A. , NASCIMENTO , M.D.D. and GOMES ,R.A.R
المصدر:
MICROBIOLOGICAL EXAMINATION METHODS OF FOOD AND WATE A Laboratory Manual
الجزء والصفحة:
10-3-2016
6476
Presence/absence method MLG/FSIS/USDA 2011 for Salmonella in foods
Method of the United States Department of Agriculture (USDA), as described in 4.05, January/2011 revision of the Microbiology Laboratory Guidebook (MLG/FSIS/USDA, 2011). It is applicable to meat, poultry and catfish products, sponge and rinse samples, and egg products. It is not intended for the isolation and identification of Salmonella Typhi.
1. Material required for analysis
• Buffered Peptone Water (BPW)
• Buffered Peptone Water (BPW) with crystal violet
(for fermented products analysis)
• Calcium carbonate (CaCO3) sterile (for fermented
products analysis)
• Rappaport-Vassiliadis (R-10) Broth or Rappaport-Vassiliadis Soya (RVS) Broth
• Tetrathionate Broth Hajna and Damon (TTH)
• Brilliant Green Sulfa (BGS) Agar
• Xylose Lysine Tergitol 4 (XLT4) Agar or Double Modified Lysine Iron Agar (DMLIA)
• Triple Sugar Iron Agar (TSI)
• Lysine Iron Agar (LIA)
• Formalinized Physiological Saline Solution
• Salmonella polyvalent flagellar (H) antiserum
• Salmonella Spicer-Edwards flagellar (H) antisera
• Salmonella polyvalent somatic (O) antiserum
• Miniaturized identification kit or biochemical tests media (the same media used in the method BAM/FDA 2011)
• Laboratory incubator set to 35 ± 2°C
• Laboratory incubator or water bath set to 42 ± 0.5°C
• Water bath set to 48–50°C
2. Procedure
A general flowchart for detection of Salmonella in foods using the presence/absence method MLG/FSIS/USDA 2011 is shown in Figure 1.
Method MLG/FSIS/USDA 2011 safety precautions: Follow CDC guidelines for Biosafety Level 2 pathogens when using live cultures of Salmonella and work in a class II laminar flow biosafety cabinet.
a) Pre-enrichment
a.1) Presence/absence test: homogenize m ± 2% grams of the test sample with 9m ± 2% milliliters of Buffered Peptone Water (BPW) (10−1 dilution). The sample analytical unity varies according the food, as described in Table 1. Incubate at 35 ± 2°C for the time established in the Table 1.
Note a.1.1) For chicken carcasses, transfer the carcass to a sterile plastic bag and weigh. Add to the bag and to the body cavity of the carcass 400 ml of BPW. Holding the carcass with one hand and closing the mouth of the bag with the other, agitate the liquid inside the bag with shaking and rotating movements, in a way so as to wash the entire internal and external surface of the carcass. Transfer the rinse fluid to a sterile container. For Salmonella analysis inoculate 30 ± 0.6 ml of the rinse fluid into 30 ± 0.6 ml of BPW. For analyses other than Salmonella prepare dilutions directly from the rinse fluid. Alternatively, the carcass may be rinsed in Butterfield’s Phosphate Buffer instead of BPW. In this case, add 30 ± 0.6 ml of double concentration BPW to 30 ± 0.6 ml of carcass-rinse fluid and mix well.
Note a.1.2) For raw meat cuts (not grounded) the washing procedure described for chicken carcasses may be used. Rinse the samples with BPW in the quantity required for a 10−1 dilution. Alternatively, mince the meat with scissors and homogenize with BPW by shaking, stomaching or blending.
Note a.1.3) For carcass sponge samples, add 50 ± 1 ml of BPW to the bag containing the sponge to bring the total volume to 60 ml.
Note a.1.4) For fermented products, use BPW with crystal violet (BPW supplemented with 1 ml/l of a 1% aqueous solution of crystal violet) and homogenize the sample with calcium carbonate sterile powder (10 g for 325 g of sample = 0.77 g for 25 g of sample).
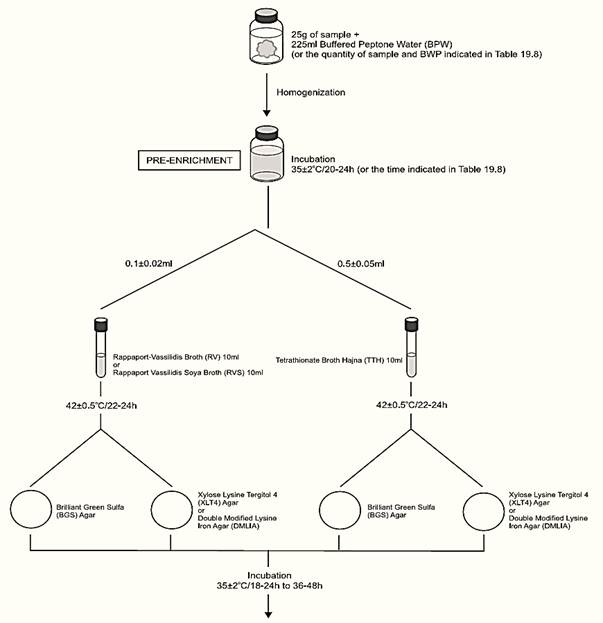
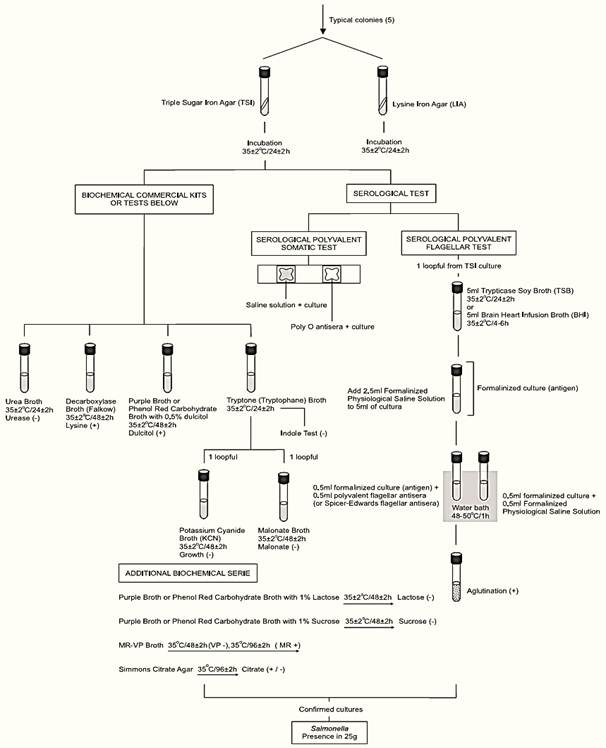
Figure 1 Scheme of analysis for detection of Salmonella in foods using the presence/absence method MLG/FSIS/USDA 2011.
Table 1 Guide for sample preparation and Salmonella pre-enrichment for cultural or PCR methods MLG/FSIS/USDA 2011
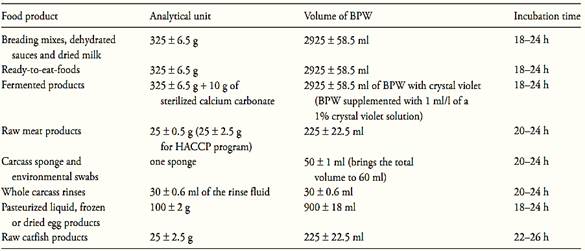
Note a.1.5) for ready-to-eat-foods containing the meat or poultry component separated from the non-meat ingredients, the FSIS analyzes only the representative meat/poultry portion. When the meat/poultry is combined with other ingredients to form the product, the combined ingredients are analyzed together.
a.2) Most probable number (MPN) count: Prepare the 1:10 (10−1) dilution as described above (item a.1). From the 10−1 dilution prepare serial decimal dilutions and inoculate three 10 ml aliquots of the 10−1 dilution into three empty sterile tubes, three 1 ml aliquots of the 10−1 dilution into three tubes with 9 ml of BPW, and three 1 ml aliquots of the 10−2 dilution into three tubes with 9 ml of BPW. Incubate the tubes and the remainder 10−1 dilution at 35 ± 2°C for the time established in the Table 1. From this point of the procedure continue the analysis separately for each tube and for the remainder 10−1 dilution.
Note a.2.1) This procedure is described in the MLG/FSIS Appendix 2.03 - Most Probable Number Procedure and Tables (MLG/FSIS/USDA, 2008). The aliquots used above (1, 0.1, and 0.01 g) are recommended for foods likely to contain a small Salmonella population (<10/g). For samples with expected count above this level, higher dilutions should be inoculated.
b) Selective enrichment: After the incubation period inoculate 0.1 ± 0.02 ml of the pre-enrichment broth into 10 ml tubes of Rappaport-Vassiliadis (R-10) Broth or Rappaport-Vassiliadis Soya (RVS) Broth and 0.5 ± 0.05 ml into 10 ml tubes of Tetrathionate Broth Hajna (TTH). Incubate the tubes at 42 ± 0.5°C for 22–24 h (laboratory incubator) or 18–24 h (water bath).
c) Selective-differential plating: From the culture obtained in TTH streak a loopful (10 μl) onto a plate of Brilliant Green Sulfa (BGS) Agar and a loopful (10 μl) onto a plate of Xylose Lysine Tergitol 4 (XLT4) Agar or Double Modified Lysine Iron Agar (DMLIA). Repeat the same procedure with the culture obtained in R-10 or RVS. Incubate the plates (inverted) at 35 ± 2°C/18–24 h and verify the presence of typical colonies. Reincubate all plates and repeat examination after 48 ± 2 h of incubation.
Typical Salmonella colonies on selective-differentia media: BGS: On BGS the Salmonella colonies are pink (lactose not fermented) and surrounded by a red color in the medium.
XLT4: On XLT4 the Salmonella colonies are red (lactose not fermented) and the center may be black (H2S produced) or not (H2S not produced). Some strains of Salmonella produce large amounts of H2S resulting in colonies completely black. The margin of the colony may be yellow in 24 h but become red with prolongation of incubation.
DMLIA: On DMLIA the Salmonella colonies are purple with or without black centers (lysine decarboxylated, lactose and sucrose not fermented).
d) Screening: From each selective agar plate select three (or more) typical colonies for confirmation. If the typical colonies on a plate are not well isolated, purify the culture using the same medium or, alternatively, inoculate a loopful of into a tube of TTH or R-10 or RVS broth, incubate at 35 ± 2°C overnight and re-streak to selective agars.
From each selected colony inoculate a tube of Triple Sugar Iron Agar (TSI) by streaking the slant and stabbing the butt. With the same inoculum, without flaming the needle, inoculate a tube of Lysine Iron Agar (LIA) by streaking the slant and stabbing the butt. Incubate the TSI and LIA tubes at 35 ± 2°C/24 ± 2 h (loose the caps to maintain aerobic conditions). After inoculation maintain the BGS and XLT4 or DMLIA plates under refrigeration (2–8°C).
Note d.1) Since lysine decarboxylation reaction is strictly anaerobic, the LIA slants must have deep butt (4 cm).
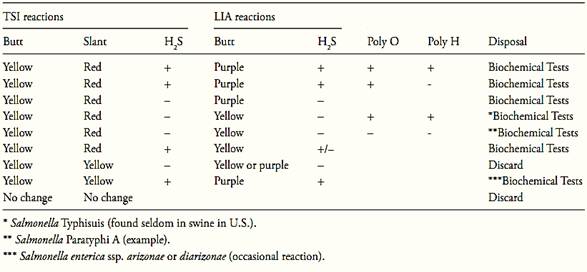
After the incubation period, examine the tubes for typical Salmonella reactions. In TSI: alkaline (red) slant and acid (yellow) butt, with or without production of H2S (blackening of agar). In LIA: alkaline (purple) reaction in butt of tube with or without production of H2S.
Discard any pair of tubes that show “swarming” from the original site of inoculation. Dis-card any pair of tubes that show a reddish slant in LIA.
Use the inoculum from the TSI or LIA slant to perform serological tests, using the same procedure described by method BAM/FDA 2011. Test first with polyvalent O antiserum (groups A to I), including a saline control with each isolate. Cultures showing agglutination with the saline control alone (auto-agglutination) should be identified by biochemical tests only. Cultures poly O positive and not showing auto-agglutination should be submitted to the serological polyvalent flagellar (H) test.
The Oxoid Salmonella Latex Test or the SSI H Antisera for Slide Agglutination, or other equivalent commercial kit may be used for the serological polyvalent flagellar (H) test (follow the manufacturer’s instructions). However, if a suspect Salmonella isolate gives a negative result by the agglutination test, the tube serological polyvalent flagellar (H) test described by method BAM/FDA 2011 should be performed.
Follow criteria from Table 2 to select cultures for biochemical tests.
e) Confirmation: Commercially available biochemical test kits, including automated systems may be used for biochemical identification, following the manufacturers’ instructions. Alternatively, use traditional methods described in the AOAC Official Method 967.27 (the same used in the BAM/FDA, 2011 method above).
Table 1 Guide for selecting TSI and LIA cultures for confirmation tests according the method MLG/FSIS/USDA 2011.
References
Silva, N.D .; Taniwaki, M.H. ; Junqueira, V.C.A.; Silveira, N.F.A. , Nasdcimento , M.D.D. and Gomes ,R.A.R .(2013) . Microbiological examination methods of food and water a laboratory Manual. Institute of Food Technology – ITAL, Campinas, SP, Brazil .
MLG/FSIS/USDA (2011) Isolation and Identification of Salmonella from Meat, Poultry, Pasteurized Egg and Catfish Products. In: Microbiology Laboratory Guidebook [Online] Washington, Food Safety and Inspection Service, United States Department of Agriculture. Available from: http://www.fsis.usda.gov/PDF/MLG_4_05.pdf [Accessed 10th February 2012].
 الاكثر قراءة في البكتيريا
الاكثر قراءة في البكتيريا
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















