

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Restriction Fragment Length Polymorphism : DNA variations resulting in RFLP
المؤلف:
Denise R. Ferrier
المصدر:
Lippincott Illustrated Reviews: Biochemistry
الجزء والصفحة:
3-1-2022
1712
Restriction Fragment Length Polymorphism : DNA variations resulting in RFLP
It has been estimated that the genomes of any two unrelated people are 99.5% identical. With 6 billion bp in the diploid human genome, that represents variation in ~30 million bp. These genome variations are the result of mutations that lead to polymorphisms. A polymorphism is a change in genotype that can result in no change in phenotype or a change in phenotype that is harmless, causes increased susceptibility to a disease, or, rarely, causes the disease. It is traditionally defined as a sequence variation at a given locus (allele) in >1% of a population. Polymorphisms primarily occur in the 98% of the genome that does not encode proteins (that is, in introns and intergenic regions).
A restriction fragment length polymorphism (RFLP) is a genetic variant that can be observed by cleaving the DNA into fragments (restriction fragments) with a restriction endonuclease. The length of the restriction fragments is altered if the variant alters the DNA so as to create or abolish a restriction site. RFLP can be used to detect human genetic variations, for example, in prospective parents or in fetal tissue.
DNA variations resulting in RFLP
Two types of DNA variations commonly result in RFLP: single-base changes in the DNA sequence and tandem repeats of DNA sequences.
1. Single-base changes: About 90% of human genome variation comes in the form of single nucleotide polymorphisms (SNPs, pronounced “snips”), that is, variations that involve just one base (Fig. 1). The substitution of one nucleotide at a restriction site can render the site unrecognizable by a particular restriction endonuclease. A new restriction site can also be created by the same mechanism. In either case, cleavage with an endonuclease results in fragments of lengths that differ from the normal and can be detected by DNA hybridization . The altered restriction site can be either at the site of a diseasecausing mutation (rare) or at a site some distance from the mutation. [Note: The HapMap, developed by The International Haplotype Map Project, is a catalog of common SNP in the human genome. The data are being used in genome-wide association studies (GWAS) to identify those alleles that affect health and disease.]
Figure 1: Common forms of genetic polymorphism. SNP = single nucleotide polymorphism; A = adenine; C = cytosine; G = guanine; T = thymine.
2. Tandem repeats: Polymorphisms in chromosomal DNA can also arise from the presence of a variable number of tandem repeats (VNTR), as shown in Figure 2. These are short sequences of DNA at scattered locations in the genome, repeated in tandem (one after another). The number of these repeat units varies from person to person but is unique for any given individual and, therefore, serves as a molecular “fingerprint.” Cleavage by restriction enzymes yields fragments that vary in length depending on how many repeated segments are contained in the fragment (see Fig. 2). Many different VNTR loci have been identified and are extremely useful for DNA fingerprint analysis, such as in forensic and paternity cases. It is important to emphasize that these polymorphisms, whether SNP or VNTR, are simply markers, which, in most cases, have no known effect on the structure, function, or rate of production of any particular protein.
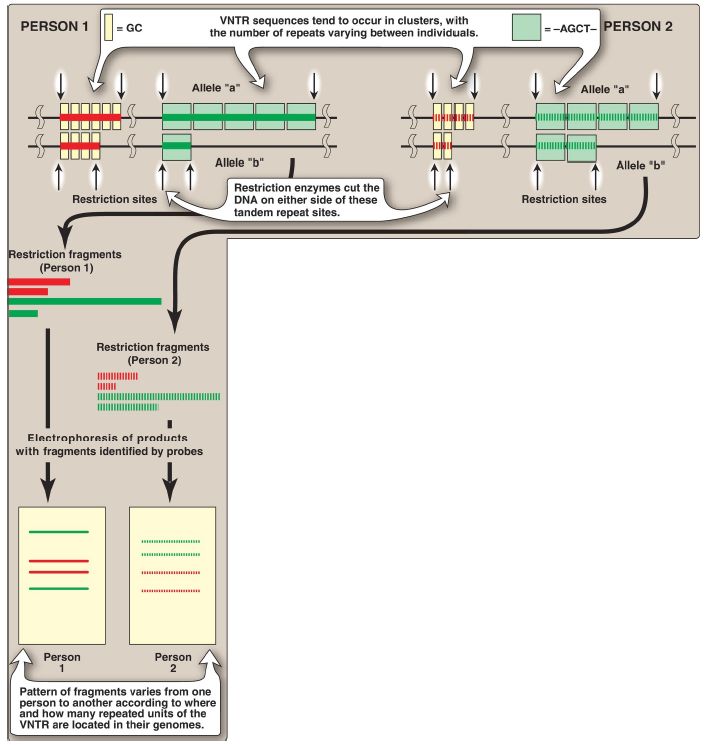
Figure 2: Restriction fragment length polymorphism of variable number tandem repeats (VNTR). For each person, a pair of homologous chromosomes is shown.
 الاكثر قراءة في الكيمياء الحيوية
الاكثر قراءة في الكيمياء الحيوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















