
Methylation of Histones and DNA Is Connected
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 11-6-2021
11-6-2021
 2429
2429
Methylation of Histones and DNA Is Connected
KEY CONCEPTS
- Methylation of both DNA and specific sites on histones is a feature of inactive chromatin.
- The SET domain is part of the catalytic site of protein methyltransferases.
- The two types of methylation event are connected.
DNA methylation is associated with transcriptional inactivity, whereas histone methylation can be linked to either active or inactive regions, depending on the specific site of methylation. Numerous sites of lysine methylation are present in the tail and core of histone H3 (a few of which occur only in some species), and a single lysine in the tail of H4 is methylated. In addition, three arginines in H3 and one in H4 are also methylated. Because lysines can be mono-, di-, or trimethylated, and arginines can be mono- or dimethylated (see the Chromatin chapter), the number of potential functional methylation marks is large.
For example, di- or trimethylation of H3K4 is associated with transcriptional activation, and trimethylated H3K4 occurs around the start sites of active genes. In contrast, H3 methylated at K9 or K27 is a feature of transcriptionally silent regions of chromatin, including heterochromatin and smaller regions containing one or more silent genes. Whole-genome studies can help to uncover general patterns of modifications linked to different transcriptional states, as shown in FIGURE 1.
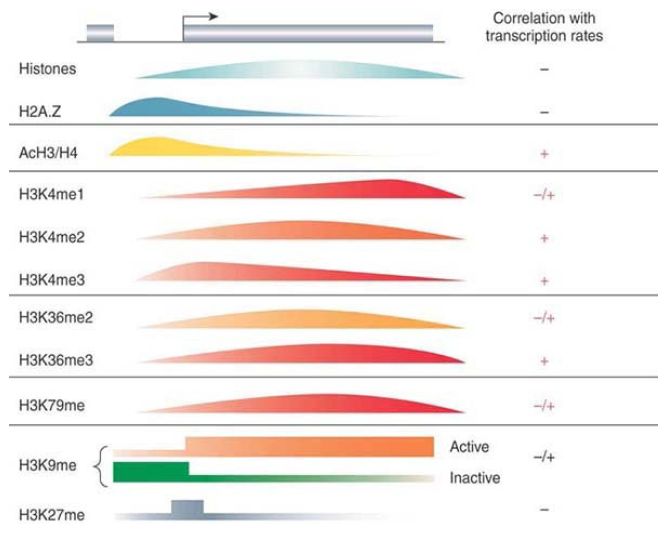
FIGURE 1. The distribution of histones and their modifications are mapped on an arbitrary gene relative to its promoter. The curves represent the patterns that are determined via genome-wide approaches. The location of the histone variant H2A.Z is also shown. With the exception of the data on K9 and K27 methylation, most of the data are based on yeast genes.
Reprinted from Cell, vol. 128, B. Li, M. Carey, and J. L. Workman, The Role of Chromatin
during Transcription, pp. 707–719. Copyright 2007, with permission from Elsevier
[http://www.sciencedirect.com/science/journal/00928674].
Histone lysine methylation is catalyzed by lysine methyltransferases (HMTs or KMTs), most of which contain a conserved region called the SET domain. Like acetylation, methylation is reversible, and two different families of lysine demethylases (KDMs) have been identified: the LSD1 (lysine-specific demethylase 1, also known as KDM1) family and the Jumonji family. Different classes of enzymes demethylate arginines.
In silent or heterochromatic regions, the methylation of H3 at K9 is linked to DNA methylation. The enzyme that targets this lysine is a SET domain–containing enzyme called Suv39h1. Deacetylation of H3K9 by HDACs must occur before this lysine can be methylated.
H3K9 methylation then recruits the protein HP1 (heterochromatin protein 1), which binds H3K9me via its chromodomain. HP1 then targets the activity of DNA methyltransferases (DNMTs). Most of the methylation sites in DNA are CpG islands (see the chapter titled Epigenetics I). CpG sequences in heterochromatin are typically methylated. Conversely, it is necessary for the CpG islands located in promoter regions to be unmethylated in order for a gene to be expressed.
Methylation of DNA and methylation of histones are connected in a mutually reinforcing circuit. In addition to the recruitment of DNMTs via HP1 binding to H3K4me, DNA methylation can, in turn, result in histone methylation. Some histone methyltransferase complexes (as well as some HDAC complexes) contain binding domains that recognize the methylated CpG doublet, thus the DNA methylationreinforces the circuit by providing a target for the histone deacetylases and methyltransferases to bind. The important point is that one type of modification can be the trigger for another.
These systems are widespread, as can be seen by evidence for these connections in fungi, plants, and animal cells, and for regulating transcription at promoters used by both RNA polymerases I and II, as well as maintaining heterochromatin in an inert state.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


