

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Mechanism of Action of Activators and Repressors
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
9-6-2021
2642
Mechanism of Action of Activators and Repressors
Key concept
- Activators determine the frequency of transcription.
- Activators work by making protein–protein contacts with the basal factors.
- Activators may work via coactivators.
- Activators are regulated in many different ways.
- Some components of the transcriptional apparatus work by changing chromatin structure.
- Repression is achieved by affecting chromatin structure or by binding to and masking activators.
Initiation of transcription involves many protein–protein interactions between transcription factors bound at enhancers with the basal apparatus that assembles at the promoter, including RNA polymerase. These transcription factors can be divided into two opposing classes: positive activators and negative repressors. As discussed in the chapter titled The Operon, positive control in bacteria entails a regulator that aids the RNA polymerase in the transition from the closed complex to the open complex. Transcription factors, such as CRP (catabolite repressor protein), in Escherichia coli, typically bind close to the promoter to allow the C-terminal domain of the α subunit of RNA polymerase to make direct physical contact. This usually occurs in a gene having a poor promoter sequence. The activator functions to overcome the inability of the RNA polymerase to open the promoter. Positive control in eukaryotes is quite different. Three classes of activators can be identified that differ by function.
The first class is the true activators . These are the classical transcription factors that function by making direct physical contact with the basal apparatus at the promoter either directly or indirectly, through a coactivator. These transcription factors function on DNA or chromatin templates.
The activity of a true activator may be regulated in any one of several ways, as illustrated schematically in FIGURE 1:
- A factor is tissue specific because it is synthesized only in a particular type of cell. This is typical of factors that regulate development, such as homeodomain proteins.
- The activity of a factor may be directly controlled by modification. HSF (heat shock transcription factor) is converted to the active form by phosphorylation.
- A factor is activated or inactivated by binding a ligand. The steroid receptors are prime examples. Ligand binding may influence the localization of the protein (causing transport from
cytoplasm to nucleus), as well as determine its ability to bind to DNA.
- Availability of a factor may vary; for example, the factor NF-κB (which activates immunoglobulin κ genes in B lymphocytes) is present in many cell types. It is sequestered or masked in the cytoplasm, however, by the inhibitory protein I-κB. In B lymphocytes, NF-κB is released from I-κB and moves to the nucleus, where it activates transcription.
- A dimeric factor may have alternative partners. One partner may cause it to be inactive; synthesis of the active partner may displace the inactive partner. Such situations may be amplified into networks in which various alternative partners pair with one another, especially among the helix-loop-helix (HLH) proteins.
- The factor may be cleaved from an inactive precursor. One activator is produced as a protein bound to the nuclear envelope and endoplasmic reticulum. The absence of sterols (such as cholesterol) causes the cytosolic domain to be cleaved; it then translocates to the nucleus and provides the active form of the activator.
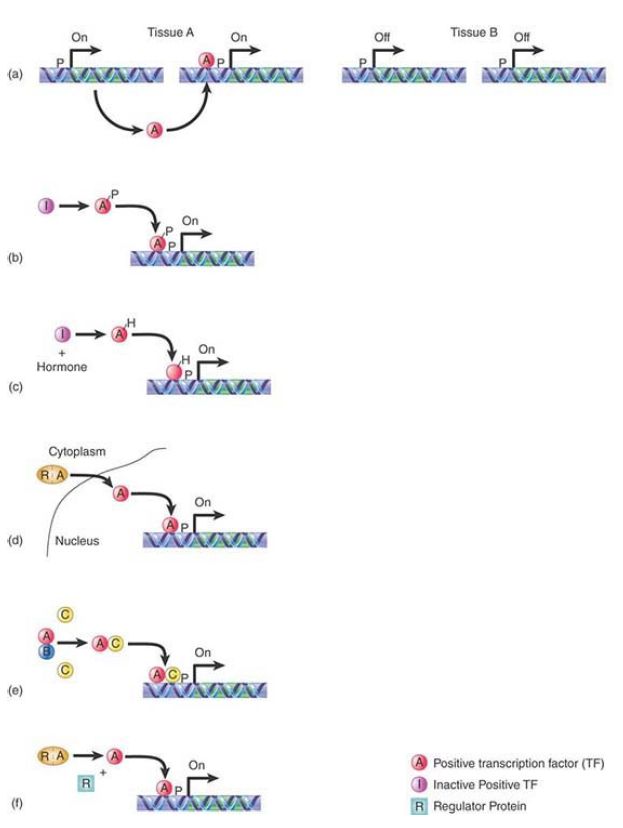
FIGURE 26.3 The activity of a positive regulatory transcription factor may be controlled by (a) synthesis of protein, (b) covalent modification of protein, (c) ligand binding, or (d) binding of inhibitors that sequester the protein or affect its ability to bind to DNA (e) by the ability to select the correct binding partner for activation and (f) by cleavage from an inactive precursor.
The second class includes the antirepressors. When one of these activators is bound to its enhancer, it recruits the histone modifier enzymes and/or the chromatin remodeler complexes to convert the chromatin from the closed state to the open state. This class has no activity on a DNA template; it only functions on chromatin templates .
The third class includes architectural proteins, such as Yin-Yang; these proteins function to bend the DNA, either bringing bound proteins together to facilitate forming a cooperative complex or bending the DNA the other way to prevent complex formation, as shown in FIGURE 2. Note that a strand of DNA may thus be bent in two different directions depending on whether the regulator binds to the top or to the bottom. This is a difference of one-half of a turn of the helix, which is about 5 bp (10.5 bp per turn).
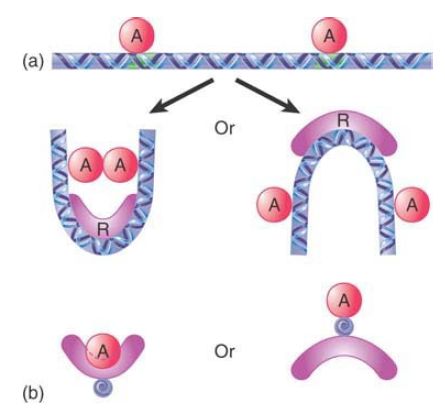
FIGURE 2.Architectural proteins control the structure of DNA and thus control whether bound proteins can contact each other.
Several examples of negative control in bacteria, in the lac operon and in the trp operon, were described in the chapter titled The Operon. Repression can occur in bacteria when the repressor prevents the RNA polymerase from converting the promoter from the closed complex to the open complex, as in the lac operon, or bind to the promoter sequence to prevent RNA polymerase from binding, as in the trp operon. Many more mechanisms have been identified by which repressors act in eukaryotes, some of which are illustrated in FIGURE 3:
- One mechanism of action by which a eukaryotic repressor can prevent gene expression is to sequester an activator in the cytoplasm. Eukaryotic proteins are synthesized in the cytoplasm. Proteins that function in the nucleus have a domain that directs their transport through the nuclear membrane. A repressor can bind to that domain and mask it.
- Several variations of that mechanism are possible. One that takes place in the nucleus occurs when the repressor binds to an activator that is already bound to an enhancer and masks its activation domain, thus preventing it from functioning, such as with the Gal80 repressor .
- Alternatively, the repressor can be masked and held in the cytoplasm until it is released to enter the nucleus.
- A fourth mechanism is simple competition for an enhancer, where either the repressor and activator have the same binding site sequence or have overlapping but different binding site sequences. This is a very versatile mechanism for a cell because there are two variables at work here: One is strength of a factor binding to DNA, and the second is factor concentration. By only slightly varying the concentration of a factor, a cell can dramatically alter its developmental path.
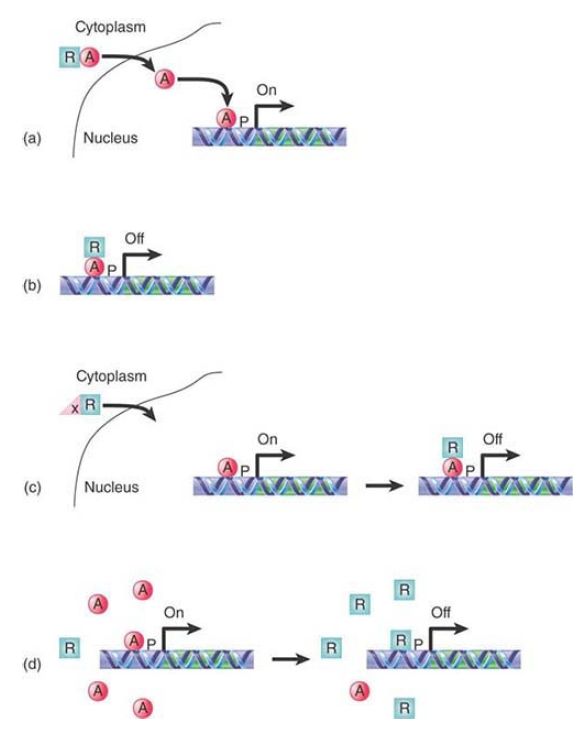
FIGURE 3. A repressor may control transcription by (a) sequestering an activator in the cytoplasm, (b) by binding an activator and masking its activation domain, (c) by being held in the cytoplasm until it is needed, or (d) by competing with an activator for a binding site.
The transcription factors that recruit the histone modifiers and chromatin remodelers have as their counterparts repressors that recruit the complexes that undo (or change) the modifications and remodeling. The same is true for the architectural proteins, where, in fact, the same protein bound to a different site prevents activator complexes from forming.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















