

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Cycle of Bacterial Messenger RNA
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
28-5-2021
3125
The Cycle of Bacterial Messenger RNA
KEY CONCEPTS
- Transcription and translation occur simultaneously in bacteria (called coupled transcription/translation) as ribosomes begin translating an mRNA before its synthesis has been completed.
- Bacterial mRNA is unstable and has a half-life of only a few minutes.
- A bacterial mRNA may be polycistronic in having several coding regions that represent different cistrons.
Messenger RNA has the same function in all cells, but there are important differences in the details of the synthesis and in the structures of prokaryotic and eukaryotic mRNAs. A major difference in the production of mRNA depends on the
cellular locations where transcription and translation occur:
- In bacteria, mRNA is transcribed and translated in the single cellular compartment; the two processes are so closely linked that they occur simultaneously. Ribosomes attach to bacterial mRNA even before its transcription has been completed so the polysome is likely to still be attached to DNA. Bacterial mRNA is usually unstable and is therefore translated into polypeptides for only a few minutes. This process is called coupled transcription/translation.
- In a eukaryotic cell, synthesis and maturation of mRNA occur exclusively in the nucleus. Only after these events are completed is the mRNA exported to the cytoplasm, where it is translated by ribosomes. A typical eukaryotic mRNA is often intrinsically stable and continues to be translated for several hours, though there is a great deal of variation in the stability of specific mRNAs, in some cases due to stability or instability sequences in the 5′ or 3′ UTRs.
Figure 1. shows that transcription and translation are intimately related in bacteria. Transcription begins when the enzyme RNA polymerase binds to DNA and then moves along, making a copy of one strand. Soon after transcription begins, ribosomes attach to the 5′ end of the mRNA and start translation, even before the rest of the mRNA has been synthesized. Multiple ribosomes move along the mRNA while it is being synthesized. The 3′ end of the mRNA is generated when transcription terminates. Ribosomes continue to translate the mRNA while it persists, but it is degraded in the overall 5′ to 3′ direction quite rapidly. The mRNA is synthesized, translated by the ribosomes, and degraded, all in rapid succession. An individual molecule of mRNA persists for only a matter of minutes at most.
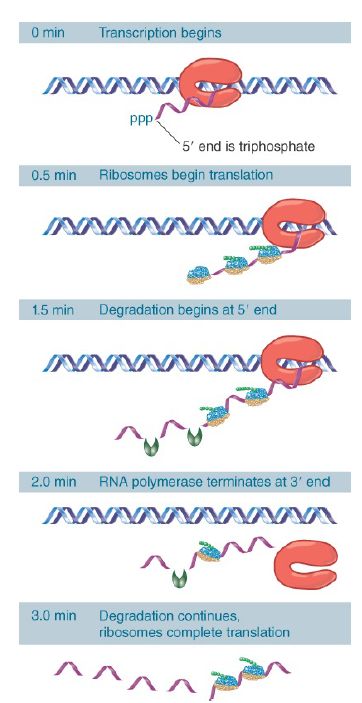
FIGURE 22.51 mRNA is transcribed, translated, and degraded simultaneously in bacteria.
Bacterial transcription and translation take place at similar rates. At 37°C, transcription of mRNA occurs at a rate of about 40 to 50 nucleotides per second. This is very close to the rate of polypeptide synthesis, which is roughly 15 amino acids per second.
It therefore takes about 1 minute to transcribe and translate an mRNA of 2,500 nucleotides, corresponding to a 90-kD polypeptide. When expression of a new gene is initiated, its mRNA will typically appear in the cell within about 1.5 minutes. The corresponding polypeptide will appear within another 30 seconds. Bacterial translation is very efficient, and most mRNAs are translated by a large number of tightly packed ribosomes. In one example, trp mRNA, about 15 initiations of transcription occur every minute and each of the 15 mRNAs is probably translated by about 30 ribosomes in the interval between its transcription and degradation.
The instability of most bacterial mRNAs is striking. Degradation of mRNA closely follows its translation and likely begins within 1 minute of the start of transcription. The 5′ end of the mRNA starts to decay before the 3′ end has been synthesized or translated.
Degradation seems to follow the last ribosome of the convoy along the mRNA. However, degradation proceeds more slowly, probably at about half the speed of transcription or translation. The stability of mRNA has a major influence on the amount of polypeptide that is produced. It is usually expressed in terms of the half-life. The mRNA representing any particular gene has a characteristic half-life, but the average is about 2 minutes in bacteria.
Of course, this series of events is only possible because transcription, translation, and degradation all occur in the same direction. The dynamics of gene expression have been “caught in the act” in the electron micrograph of Figure 1 . In these (unknown) transcription units, several mRNAs are undergoing synthesis simultaneously, and each carries many ribosomes engaged in translation. An RNA whose synthesis has not yet been completed is called a nascent RNA.
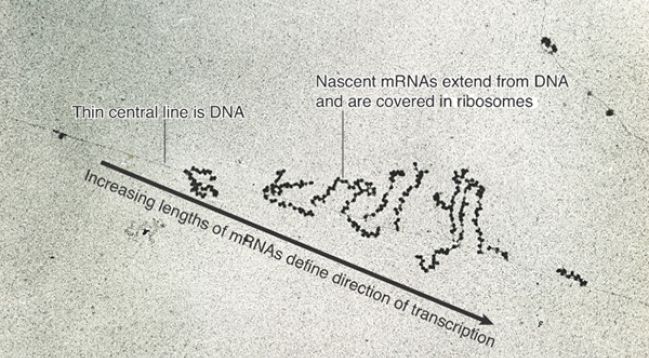
FIGURE 22.52 Transcription units can be visualized in bacteria.
© Prof. Oscar L. Miller/Photo Researchers, Inc.
Bacterial mRNAs vary greatly in the number of proteins that they encode. Some mRNAs carry only a single ORF; they are monocistronic. Others (the majority) carry sequences encoding several polypeptides; they are polycistronic. In these cases, a single mRNA is transcribed from a group of adjacent cistrons. (Such a cluster of cistrons constitutes an operon that is controlled as a single genetic unit; see The Operon chapter.)
All mRNAs contain three regions. The coding region, or open reading frame (ORF), consists of a series of codons representing the amino acid sequence of the polypeptide, starting (usually) with AUG and ending with one of the three termination codons.
However, the mRNA is always longer than the coding region as extra regions are present at both ends. An additional sequence at the 5′ end, upstream of the coding region, is described as the leader or 5′ UTR. An additional sequence downstream from the termination signal, forming the 3′ end, is called the trailer or 3′ UTR. Although they do not encode a polypeptide, these sequences may contain important regulatory instructions, especially in eukaryotic mRNAs.
A polycistronic mRNA also contains intercistronic regions, as illustrated in Figure 2 . They vary greatly in size. They may be as long as 30 nucleotides in bacterial mRNAs (and even longer in phage RNAs), or they may be very short, with as few as one or two nucleotides separating the termination codon for one polypeptide from the initiation codon for the next. In an extreme case, two genes actually overlap, so that the last base of one coding region is also the first base of the next coding region.
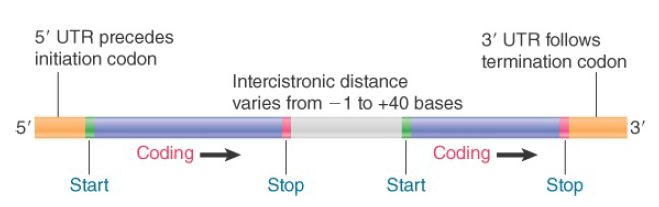
FIGURE 2. Bacterial mRNA includes untranslated as well as translated regions. Each coding region has its own initiation and termination signals. A typical mRNA may have several coding regions (ORFs).
The number of ribosomes engaged in translating a particular cistron depends on the efficiency of its initiation site in the 5′ UTR. The initiation site for the first cistron becomes available as soon as the 5′ end of the mRNA is synthesized. How are subsequent cistrons translated? Are the several coding regions in a polycistronic mRNA translated independently, or is their expression connected? Is the mechanism of initiation the same for all cistrons, or is it different for the first cistron and the downstream cistrons?
Translation of a bacterial mRNA proceeds sequentially through its cistrons. At the time when ribosomes attach to the first coding region, the subsequent coding regions have not yet been transcribed. By the time the second ribosomal binding site is available, translation is well under way through the first cistron.
Typically, ribosomes terminate translation at the end of each cistron, and then a new ribosome assembles independently at the start of the next coding region. This is influenced by the intercistronic region and the density of ribosomes on the mRNA.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















