

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Other Degradation Pathways Target Specific mRNAs
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
19-5-2021
2532
Other Degradation Pathways Target Specific mRNAs
KEY CONCEPTS
- Four additional degradation pathways involve regulated degradation of specific mRNAs.
- Deadenylation-independent decapping proceeds in the presence of a long poly(A) tail.
- The degradation of the nonpolyadenylated histone mRNAs is initiated by 3′ addition of a poly(U) tail.
- Degradation of some mRNAs may be initiated by sequence- or structure-specific endonucleolytic cleavage.
- An unknown number of mRNAs are targeted for degradation or translational repression by microRNAs.
Four other pathways for mRNA degradation have been described. FIGURE 1and TABLE .1 summarize these, along with the two major pathways. These pathways are specific for subsets of mRNAs and typically involve regulated degradation events.
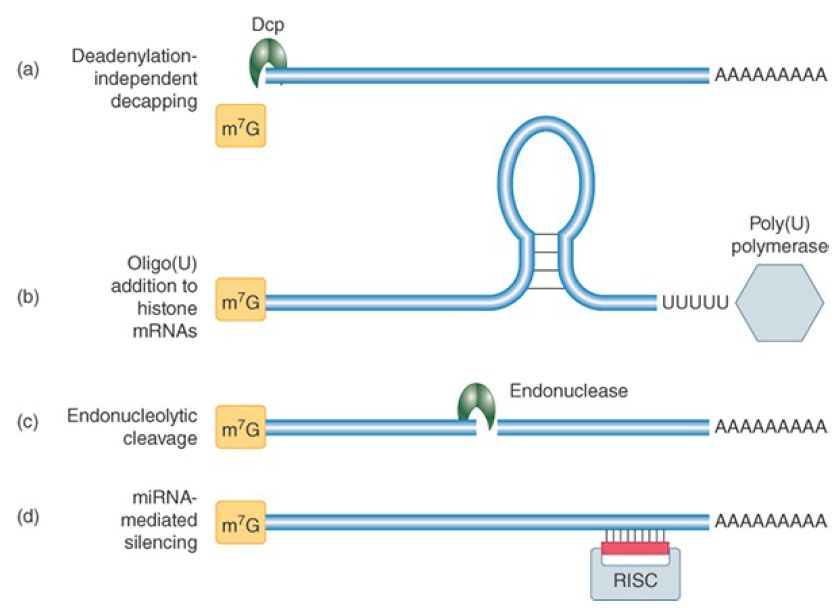
FIGURE 1 Other decay pathways in eukaryotic cells. The initiating event for each pathway is illustrated. (a) Some mRNAs may be decapped before deadenylation occurs. (b) Histone mRNAs receive a short poly(U) tail to become a decay substrate. (c) Degradation of some mRNAs can be initiated by a sequencespecific endonucleolytic cut. (d) Some mRNAs can be targeted for degradation or translational silencing by complementary guide miRNAs.
TABLE 20.1 Summary of key elements of mRNA decay pathways in eukaryotic cells.
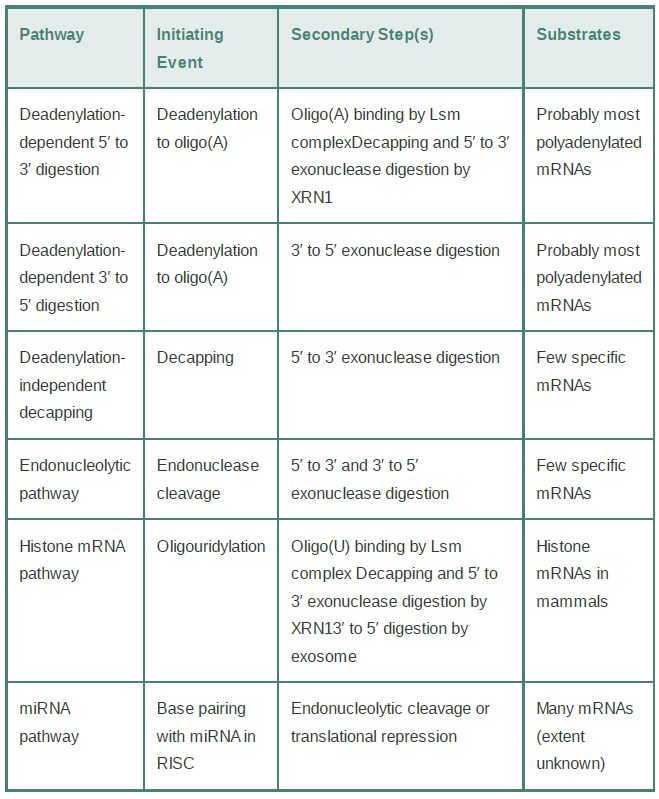
One pathway involves deadenylation-independent decapping; that is, decapping proceeds in the presence of a still long poly(A) tail. Decapping is then followed by Xrn1 digestion. Bypassing the deadenylation step requires a mechanism to recruit the decapping machinery and inhibit eIF4F binding without the help of the Lsm1–7 complex. One of the mRNAs degraded by this pathway is RPS28B mRNA, which encodes the ribosomal protein S28 and has an interesting autoregulation mechanism. A stem-loop in its 3′ UTR is involved in recruiting a known decapping enhancer. The recruitment occurs only when the stem-loop is bound by S28 protein. Thus, an excess of free S28 in the cell will cause the accelerated decay of its mRNA.
A second specialized pathway is used to degrade the cell cycle–regulated histone mRNAs in mammalian cells. These mRNAs are responsible for synthesis of the huge number of histone proteins needed during DNA replication. They accumulate only during Sphase and are rapidly degraded at its end. The nonpolyadenylated histone mRNAs terminate in a stem-loop structure similar to that of many bacterial mRNAs. Their mode of degradation has striking similarities to bacterial mRNA decay. A polymerase, structurally similar to the bacterial poly(A) polymerase, adds a short poly(U) tail instead of a poly(A) tail. This short tail serves as a platform for the Lsm1–7 complex and/or the exosome, activating the standard decay pathways. This mode of degradation provides an important evolutionary link between mRNA decay systems in prokaryotes and eukaryotes.
A third pathway is initiated by sequence- or structure-specific endonucleotic cleavage. The cleavage is followed by 5′ to 3′ and 3′ to 5′ digestion of the fragments, and a scavenging decapping enzyme, different from the Dcp complex, can remove the cap.
Several endonucleases that cleave specific target sites in mRNAs have been identified. One interesting case is the targeted cleavage of yeast CLB2 (cyclin B2) mRNA, which occurs only at the end of mitosis. The endonuclease that catalyzes the cleavage, RNase MRP, is restricted to the nucleolus and mitochondria for most of the cell cycle, where it is involved in RNA processing but is transported to the cytoplasm in late mitosis.
The fourth, and most important, pathway is the microRNA (miRNA) pathway. This pathway usually leads directly to endonucleolytic cleavage of mRNA in plants; in animal cells it directs targeted deadenylation-dependent degradation and, more commonly, translational repression. MicroRNAs are short RNAs (about 22 nucleotides) derived from transcribed miRNA genes and are generated by cleavage from longer precursor RNAs. In all cases, an mRNA is targeted for silencing by the base pairing of the short complementary miRNAs presented in the context of a protein complex called RISC (RNA-induced silencing complex). Thus, the silencing of target mRNAs is controlled by regulated transcription of the miRNA genes. The details of this mechanism are described in the Regulatory RNA chapter.
The significance of the microRNA pathway to total mRNA decay is substantial. At least 1,000 miRNAs are predicted to function in humans. By identification of conserved complementary target sites in the vertebrate transcriptome, it has been estimated that 50% of all mRNAs could be regulated by miRNAs. Potentially regulated mRNAs often contain multiple target sites in their 3′ UTRs. Mutation of miRNA target sites is likely to explain many genetic disease alleles, and dysregulation of miRNA has already been associated with hundreds of diseases.
An integrated model of mRNA degradation has been proposed. This model suggests that the deadenylation-dependent decay pathways represent the default systems for degrading all polyadenylated mRNAs. The rate of deadenylation and/or other steps in degradation by these pathways can be controlled by cisacting elements in each mRNA and trans-acting factors present in the cell. Superimposed on the default system are the mRNA decay pathways described earlier for targeting specific mRNAs.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















