

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
tRNA Splicing Involves Cutting and Rejoining in Separate Reactions
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
17-5-2021
3190
tRNA Splicing Involves Cutting and Rejoining in Separate Reactions
KEY CONCEPTS
- RNA polymerase III terminates transcription in poly(U) sequence embedded in a GC-rich sequence.
- tRNA splicing occurs by successive cleavage and ligation reactions.
- An endonuclease cleaves the tRNA precursors at both ends of the intron.
- Release of the intron generates two half-tRNAs with unusual ends that contain 5′–OH hydroxyl and 2′,3′-cyclic phosphate.
- The 5′–OH end is phosphorylated by a polynucleotide kinase, the cyclic phosphate group is opened by phosphodiesterase to generate a 2′-phosphate terminus and 3′–OH group, the exon ends are joined by an RNA ligase, and the 2′-phosphate is removed by a phosphatase.
Most splicing reactions depend on short consensus sequences and occur by transesterification reactions in which breaking and forming bonds are coordinated. The splicing of tRNA genes is achieved by a different mechanism that relies upon separate cleavage and ligation reactions.
Some 59 of the 272 nuclear tRNA genes in the yeast S. cerevisiae are interrupted. Each has a single intron that is located just one nucleotide beyond the 3′ side of the anticodon. The introns vary in length from 14 to 60 bases. Those in related tRNA genes are related in sequence, but the introns in tRNA genes representing different amino acids are unrelated. No consensus sequence exists that could be recognized by the splicing enzymes. This is also true of interrupted nuclear tRNA genes of plants, amphibians, and mammals.
All the introns include a sequence that is complementary to the anticodon of the tRNA. This creates an alternative conformation for the anticodon arm in which the anticodon is base paired to form an extension of the usual arm. An example is shown in FIGURE 1. Only the anticodon arm is affected—the rest of the molecule retains its usual structure.
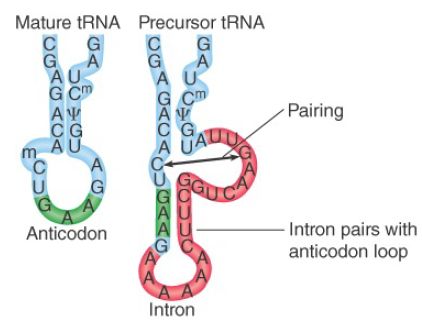
FIGURE 1. The intron in yeast tRNAPhe base pairs with the anticodon to change the structure of the anticodon arm. Pairing between an excluded base in the stem and the intron loop in the precursor may be required for splicing.
The exact sequence and size of the intron are not important. Most mutations in the intron do not prevent splicing. Splicing of tRNA depends principally on recognition of a common secondary structure in tRNA rather than a common sequence of the intron.
Regions in various parts of the molecule are important, including the stretch between the acceptor arm and D arm, in the TψC arm, and especially in the anticodon arm. This is reminiscent of the structural demands placed on tRNA for translation .
The intron is not entirely irrelevant, however. Pairing between a base in the intron loop and an unpaired base in the stem is required for splicing. Mutations at other positions that influence this pairing (e.g., to generate alternative patterns for pairing) influence splicing.
The rules that govern availability of tRNA precursors for splicing resemble the rules that govern recognition by aminoacyl-tRNA synthetases (see the chapter titled Using the Genetic Code). In a temperature-sensitive mutant of yeast that fails to remove the introns, the interrupted precursor RNAs accumulate in the nucleus. The precursors can be used as substrates for a cell-free system extracted from wild-type cells. The splicing of the precursor can be followed by virtue of the resulting size reduction of the RNA product. This is seen by the change in position of the band on gel electrophoresis, as illustrated in FIGURE 2. The reduction in size can be accounted for by the appearance of a band representing the intron.
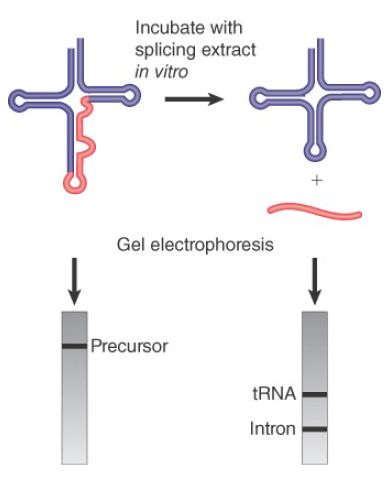
FIGURE 2.Splicing of yeast tRNA in vitro can be followed by assaying the RNA precursor and products by gel electrophoresis.
The cell-free extract can be fractionated by assaying the ability to splice the tRNA. The in vitro reaction requires ATP. Characterizing the reactions that occur with and without ATP shows that the two separate stages of the reaction are catalyzed by different enzymes:
- The first step does not require ATP. It involves phosphodiester bond cleavage by an atypical nuclease reaction. It is catalyzed by an endonuclease.
- The second step requires ATP and involves bond formation; it is a ligation reaction, and the responsible enzyme activity is described as an RNA ligase.
Splicing of pre-tRNA to remove introns is essential in all organisms, but different organisms use different mechanisms to accomplish pre-tRNA splicing. In bacteria, introns in pre-tRNAs are self-spliced as group I or group II autocatalytic introns. In archaea and eukaryotes, pre-tRNA splicing involves the action of three enzymes: (1) an endonuclease that recognizes and cleaves the precursor at both ends of the intron, (2) a ligase that joins the tRNA exons, (3) and a 2′-phosphotransferase that removes the 2′-phosphate on spliced tRNA.
The yeast endonuclease is a heterotetrameric protein consisting of two catalytic subunits, Sen34 and Sen2, and two structural subunits, Sen54 and Sen15. Its activities are illustrated in FIGURE 3. The related subunits, Sen34 and Sen2, cleave the 3′ and 5′ splice sites, respectively. Subunit Sen54 may determine the sites of cleavage by “measuring” distance from a point in the tRNA structure. This point is in the elbow of the (mature) L-shaped structure. The role of subunit Sen15 is not known, but its gene is essential in yeast. The base pair that forms between the first base in the anticodon loop and the base preceding the 3′ splice site is required for 3′ splice-site cleavage.
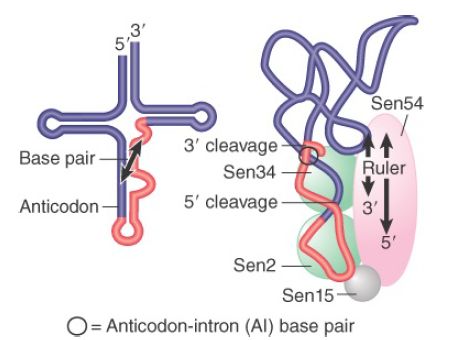
FIGURE 3.The 3′ and 5′ cleavages in S. cerevisiae pre-tRNA are catalyzed by different subunits of the endonuclease. Another subunit may determine location of the cleavage sites by measuring distance from the mature structure. The AI base pair is also important.
An interesting insight into the evolution of tRNA splicing is provided by the endonucleases of archaea. These are homodimers or homotetramers, in which each subunit has an active site (although only two of the sites function in the tetramer) that cleaves one of the splice sites. The subunit has sequences related to the sequences of the active sites in the Sen34 and Sen2 subunits of the yeast enzyme. The archaeal enzymes recognize their substrates in a different way, though. Instead of measuring distance from particular sequences, they recognize a structural feature called the bulge-helix-bulge. FIGURE 4 shows that cleavage occurs in the two bulges. Thus, the origin of splicing of tRNA precedes the separation of the archaea and the eukaryotes. If it originated by insertion of the intron into tRNAs, this must have been a very ancient event.
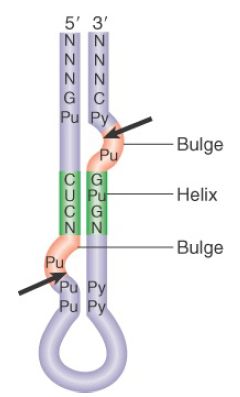
FIGURE 4. Archaeal tRNA-splicing endonuclease cleaves each strand at a bulge in a bulge-helix-bulge motif.
The overall tRNA splicing reaction is summarized in FIGURE 5. The products of cleavage are a linear intron and two half-tRNA molecules. These intermediates have unique ends. Each 5′ terminus ends in a hydroxyl group; each 3′ terminus ends in a 2′,3′- cyclic phosphate group.
The two half-tRNAs base pair to form a tRNA-like structure. When ATP is added, the second reaction occurs, which is catalyzed by a single enzyme with multiple enzymatic activities:
- Cyclic phosphodiesterase activity. Both of the unusual ends generated by the endonuclease must be altered prior to the ligation reaction. The cyclic phosphate group is first opened to generate a 2′-phosphate terminus.
- Kinase activity. The product has a 2′-phosphate group and a 3′–OH group. The 5′–OH group generated by the endonuclease must be phosphorylated to give a 5′-phosphate. This generates a site in which the 3′–OH is next to the 5′-phosphate.
- Ligase activity. Covalent integrity of the polynucleotide chain is then restored by ligase activity. The spliced molecule is now uninterrupted, with a 5′–3′ phosphate linkage at the site of splicing, but it also has a 2′-phosphate group marking the event on the spliced tRNA. In the last step, this surplus group is removed by a phosphatase, which transfers the 2′-phosphate to NDP to form ADP ribose 1′,2′-cyclic phosphate.
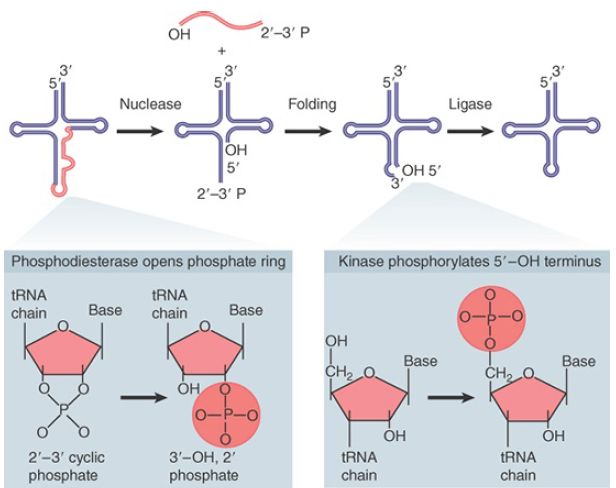
FIGURE 5.Splicing of tRNA requires separate nuclease and ligase activities. The exon–intron boundaries are cleaved by the nuclease to generate 2′,3′-cyclic phosphate and 5′–OH termini. The cyclic phosphate is opened to generate 3′–OH and 2′-phosphate groups. The 5′–OH is phosphorylated. After releasing the intron, the tRNA half molecules fold into a tRNA-like structure that now has a 3′–OH, 5′–P break. This is sealed by a ligase.
The tRNA splicing pathway described here is slightly different from that of vertebrates. Before the action of the RNA ligases, a cyclase generates a 2′,3′ cyclic terminus from the initial 3′-phosphomonoester terminus via a 3′ adenylated intermediate. The RNA ligase is also different from that in yeast because it can join a 2′,3′-cyclic phosphodiester and a 5′–OH to form a conventional 3′,5′-phosphodiester bond, but these reactions leave no extra 2′-phosphate.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















