

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Competition for Sigma Factors Can Regulate Initiation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
6-5-2021
2560
Competition for Sigma Factors Can Regulate Initiation
KEY CONCEPTS
- E. coli has seven sigma factors, each of which causes RNA polymerase to initiate at a set of promoters defined by specific −35 and −10 sequences.
- The activities of the different sigma factors are regulated by different mechanisms.
In the next few sections, we provide a few examples of regulation of initiation, elongation, and termination. Other examples will be presented in the chapters titled The Operon and Phage Strategies.
The division of labor between a core enzyme responsible for chain elongation and a sigma factor responsible for promoter selection raised the question of whether there would be more than one type of sigma factor, each specific for a different set of promoters. FIGURE 1shows the principle of a system in which a substitution of the sigma factor changes the choice of promoter.
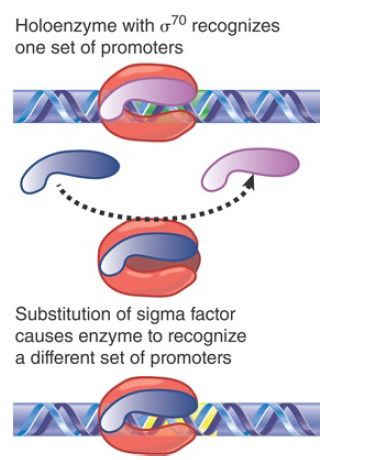
FIGURE 1. The sigma factor associated with core enzyme determines the set of promoters at which transcription is initiated.
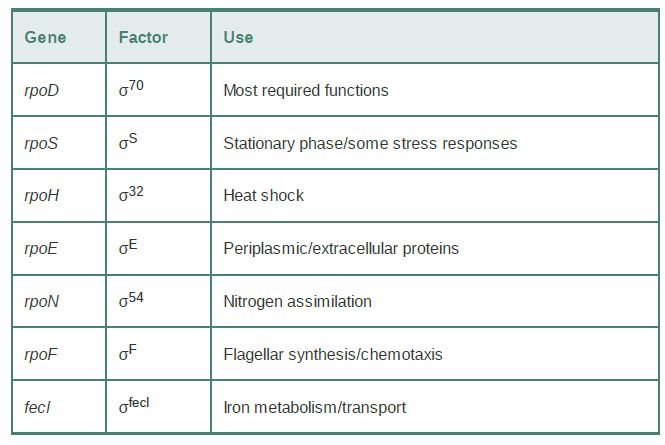
E. coli often uses alternative sigma factors to respond to changes in environmental or nutritional conditions; they are listed in TABLE 1 (sigma factors are named by the molecular weight of the product or by the function of the genes they transcribe). The most abundant sigma factor, responsible for transcription of most genes under normal conditions, is σ70 (called σA in most bacterial species) and is encoded by the rpoD gene. The alternative sigma factor σS(σ32 ) is used for making many stress-related products; σH (σ32 ) and σE (σ24 ) are required for making products needed for responding to conditions that unfold proteins in the cytoplasm and periplasm, respectively; σN (σ54 ) makes products needed primarily for nitrogen assimilation; σFecl (σ19 ) makes a few products needed for iron transport; and σF (σ28 ) expresses products needed for synthesis of flagella.
TABLE 17.2 In addition to σ70 , E. coli has several sigma factors that are induced by particular environmental conditions. (A number in the name of a factor indicates its mass.)
The unfolded protein response is one of the most conserved regulatory responses in all of biology. Originally discovered as a response to an increase in temperature (and therefore called the heat-shock response), a similar set of proteins is synthesized in all three biological kingdoms that protect cells against environmental stress. Many of these heat-shock proteins are chaperones that reduce the levels of unfolded proteins by refolding them or degrading them. In E. coli, the induction of heat-shock proteins occurs at the transcription level. The gene rpoH is a regulator needed to switch on the heat-shock response. Its product, σ32 , is an alternative sigma factor that recognizes the promoters of the heat-shock genes.
The heat-shock response (mostly chaperones and proteases) is feedback regulated. The key to the control of σ32 is that the availability of these cytoplasmic proteases and chaperones is dependent on whether they are titrated away by unfolded proteins. Thus, when unfolded protein levels go down (either because the heat-shock proteins refold or degrade them or because the temperature is lowered), they no longer titrate away the proteases that degrade σ32 , and σ32 levels return to normal. Because σ70 and σ32 compete for available core enzyme, transcription from heatshock gene promoters returns to basal levels as σ24 and σ32 levels go back to normal. Thus, the set of gene products made during heat shock depends on the balance between σ70 and σ32 . Consistent with the importance of sigma competition, the concentration of σ70 is greater than that of core RNA polymerase under σ32 noninducing conditions.
σ32 is not the only sigma factor that controls the unfolded protein response. σE is induced by accumulation of unfolded proteins in the periplasmic space and outer membrane (rather than in the cytoplasm). As with σ32 , proteolysis is the key to induction of transcription of σE -dependent promoters. The intricate circuit responsible for regulation of σ activity is summarized in FIGURE 2. σE binds to a protein (RseA) that is located in the inner membrane. RseA is an example of an antisigma factor. When bound to σE , RseA prevents σE from binding to core RNA polymerase and activating σE promoters. These promoters transcribe products needed for refolding denatured periplasmic proteins or degrading them. Thus, the periplasmic heat-shock response is a transient feedback response controlled by the concentrations of its own gene products. The σE regulon responds to the levels of unfolded and denatured periplasmic proteins rather than unfolded and denatured cytoplasmic proteins.
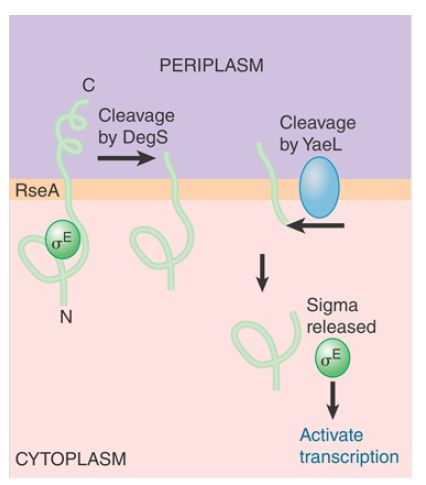
FIGURE 2 RseA is synthesized as a protein in the inner membrane. Its cytoplasmic domain binds the σE factor. RseA is cleaved sequentially in the periplasmic space and then in the cytoplasm. The cytoplasmic cleavage releases σE .
How does RseA know when to release σE ? The mechanism involves regulated, sequential proteolysis of RseA. The accumulation of unfolded proteins activates a protease (DegS) in the periplasmic space, which cleaves off the C-terminal end of the RseA protein. This cleavage activates another protease, RseP, this time on the cytoplasmic face of the inner membrane. RseP cleaves the N-terminal region of RseA, ultimately releasing σE . σE can then bind core RNA polymerase and activate transcription. Thus, accumulation of unfolded proteins at the periphery of the bacterium activates the set of genes controlled by the sigma factor.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















