

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Bacterial RNA Polymerase Consists of Multiple Subunits
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
3-5-2021
3581
Bacterial RNA Polymerase Consists of Multiple Subunits
KEY CONCEPTS
- Bacterial RNA core polymerases are multisubunit complexes of about 400 kD with the general structure αα2 ββ′ω.
- Catalysis derives from the β and β′ subunits.
The best genetically and biochemically characterized RNA polymerases are from bacteria, especially Escherichia coli. Highresolution crystal structures have been solved from two thermophilic bacterial species, Thermus aquaticus and Thermus thermophilus. Nevertheless, in all bacteria a single type of RNA polymerase is responsible for the synthesis of rRNA, mRNA, and tRNA, unlike the situation in eukaryotes where 18/28S rRNAs, mRNAs, and tRNAs typically are transcribed by different RNA polymerases (i.e., Pol I, II, and III). About 13,000 RNA polymerase molecules are present in an E. coli cell, although the precise number varies with the growth conditions. Although not all the RNA polymerases are actually engaged in transcription at any one time, almost all are bound either specifically or nonspecifically to DNA.
The complete enzyme, or holoenzyme, in E. coli has a molecular weight of about 460 kD. The holoenzyme (α2 ββ′ωσ) can be separated into two components: the core enzyme (α2 ββ′ω) and the sigma factor (the σ polypeptide), which is concerned specifically with promoter recognition. Its subunit composition is summarized in FIGURE 1. The β and β′ subunits together account for RNA catalysis and make up most of the enzyme by mass. Their amino acid sequences and their three-dimensional structures are conserved with those of the largest subunits of the RNA polymerases from all three domains of life—bacteria, archaea, and eukaryotes (see the chapter titled Eukaryotic Transcription)— indicating that the basic features of transcription are shared among the multisubunit RNA polymerases of all organisms. β and β′
together form the enzyme’s active center, the main channel through which the DNA passes during the transcription cycle, the secondary channel through which the substrate ribonucleotides enter the enzyme on their path to the active site, and the exit channel through which the nascent RNA leaves the enzyme. Consistent with the role of these subunits in all these functions, mutations in rpoB and rpoC, the genes coding for β and β′, affect all stages of transcription.
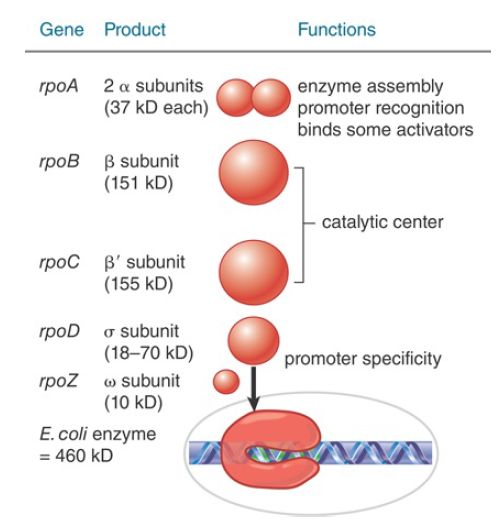
FIGURE 1. Eubacterial RNA polymerases have five types of subunits: α, β, β′, and ω have rather constant sizes in different bacterial species, but σ varies more widely.
The dimer formed by the two α subunits serves as a scaffold for assembly of the core enzyme. The C-terminal domain (CTD) of the α subunits also contacts promoter DNA directly and thereby contributes to promoter recognition .
Furthermore, the α and σ subunits are the major surfaces on RNA polymerase for interactions of the enzyme with factors that regulate transcription initiation. The ω subunit also plays a role in enzyme assembly and participates in certain regulatory functions.
The σ subunit is primarily responsible for promoter recognition. The crystal structure of the bacterial core enzyme shows that it has a crab claw–like shape, with one claw formed primarily by the β subunit and the other primarily by the β′ subunit, as illustrated in FIGURE 2. The main channel for DNA lies at the interface of the β and β′ subunits, which stabilize the separated single strands in the transcription bubble, as shown in FIGURE 3.
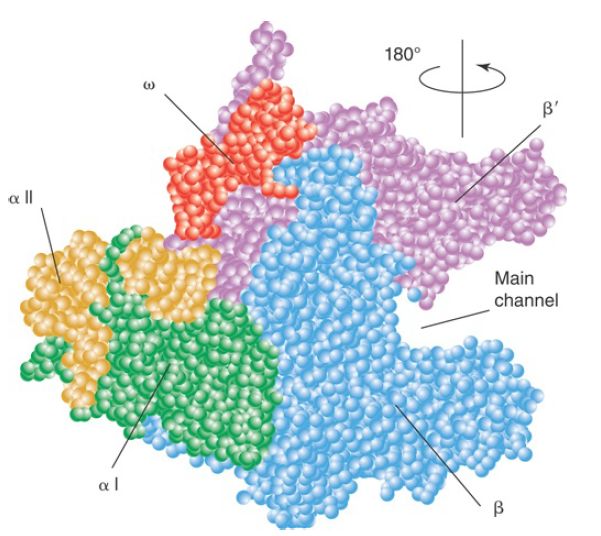
FIGURE 2. The upstream face of the core RNA polymerase, illustrating the “crab claw” shape of the enzyme. The β (cyan) and β′ (pink) subunits of RNA polymerase have a channel for the DNA template. αI is shown in green and αII in yellow; ω is red.
Data from K. M. Geszvain and R. Landick (ed. N. P. Higgins). The Bacterial Chromosome. American Society for Microbiology, 2004.
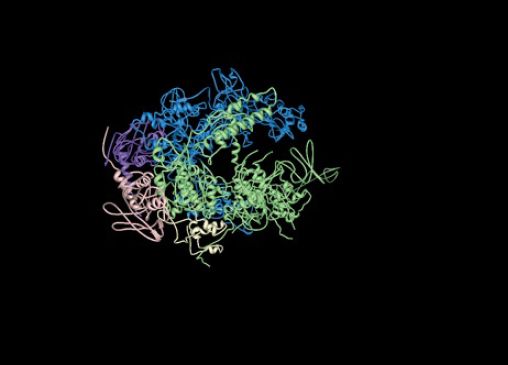
FIGURE 3. The structure of RNA polymerase core enzyme for the bacterium Thermus aquaticus, with the β subunit in blue and the β′ subunit in green.
Structure from Protein Data Bank 1HQM. L. Minakhin, et al., Proc. Natl. Acad. Sci. USA 98 (2001): 892–897.
The catalytic site is at the base of the cleft formed by the β and β′ “jaws.” One of the two catalytic Mg ions needed for the mechanism of catalysis is tightly bound to the enzyme in the active site . The other Mg arrives at the active site in a complex with the incoming nucleoside triphosphate (NTP). As indicated earlier, the eukaryotic core enzyme has the same basic structure as the bacterial enzyme, although it contains some additional subunits and sequence features not found in the bacterial enzyme. The major differences between the bacterial and eukaryotic enzymes are almost exclusively at the periphery of the enzyme, far from the active site.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















