

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Single-Copy Plasmids Have a Partitioning System
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
8-4-2021
3688
Single-Copy Plasmids Have a Partitioning System
KEY CONCEPTS
- Single-copy plasmids exist at one plasmid copy per bacterial chromosome origin.
- Multicopy plasmids exist at more than one plasmid copy per bacterial chromosome origin.
- Partition systems ensure that duplicated plasmids are segregated to different daughter cells produced by a division.
The type of system that a plasmid uses to ensure that it is distributed to both daughter cells at division depends upon its type of replication system. Each type of plasmid is maintained in its bacterial host at a characteristic copy number:
- Single-copy control systems resemble that of the bacterial chromosome and result in one replication per cell division. A single-copy plasmid effectively maintains parity with the bacterial chromosome.
- Multicopy control systems allow multiple initiation events per cell cycle, with the result that there are several copies of the plasmid per bacterium. Multicopy plasmids exist in a characteristic number (typically 10 to 20) per bacterial chromosome. Copy number is primarily a consequence of the type of replication control mechanism. The system responsible for initiating replication determines how many origins can be present in the bacterium. Each plasmid consists of a single replicon, and as a result the number of origins is the same as the number of plasmid molecules. Single-copy plasmids have a system for replication control whose consequences are similar to those of the system for replication governing the bacterial chromosome. A single origin can be replicated once, and then the daughter origins are segregated to the different daughter cells.
Multicopy plasmids have a replication system that allows a pool of origins to exist. If the number is great enough (in practice, fewer than 10 per bacterium), an active segregation system becomes unnecessary, because even a statistical distribution of plasmids to daughter cells will result in the loss of plasmids at frequencies of less than 10-6 .
Plasmids are maintained in bacterial populations with very low rates of loss (less than 10-7 per cell division is typical, even for a single-copy plasmid). The systems that control plasmid segregation can be identified by mutations that increase the frequency of loss, but that do not act upon replication itself. Several types of mechanisms are used to ensure the survival of a plasmid in a bacterial population. It is common for a plasmid to carry several systems, often of different types, all acting independently to ensure its survival. Some of these systems act indirectly, whereas others are concerned directly with regulating the partition event. In terms of evolution, however, all serve the same purpose—to help ensure perpetuation of the plasmid to the maximum number of progeny bacteria.
Single-copy plasmids require partition systems to ensure that the duplicate copies find themselves on opposite sides of the septumat cell division and are therefore segregated to a different daughter cell. In fact, functions involved in partition were first identified in plasmids. FIGURE 1. summarizes the components of a common system. Typically, there are two trans-acting loci (usually called parA and parB) and a cis-acting element (usually called parS) located next to the two genes. ParA is a partition ATPase. It binds to ParB, which binds to the parS site on DNA. Deletions of any of the three loci prevent proper partition of the plasmid.
Systems of this type have been characterized for the plasmids F, P1, and R1. Partition systems generally fall into two major classes that depend on properties of the system’s ATPase. In one group, such as the system in plasmid R1, the ATPase resembles actin and acts via polymerization (discussed further in subsequent paragraphs). The other group, which includes plasmids P1 and F, has a different type of ATPase (based on protein sequence homologies). These ParAs use the bacterial nucleoid for positioning plasmids, although the mechanisms by which this is accomplished are not yet clear.

FIGURE 1. A common segregation system consists of genes parA and parB and the target site parS.
parS plays a role for the plasmid that is equivalent to the centromere of a eukaryotic chromosome. Binding of the ParB protein to it creates a structure that segregates the plasmid copies to opposite daughter cells. In some plasmids, such as P1, a bacterial protein, IHF, also binds at this site to form part of the structure. The complex of ParB (and IHF in some cases) with parS is called the partition complex. Formation of this initial complex enables further molecules of ParB to bind cooperatively, forming a very large protein–DNA complex. These complexes hold daughter plasmids together in pairs until ready to interact with ParA. The activity of ParA is necessary to position the plasmids in the cell so that at least one copy is on each side of the dividing cell septum.
The partition ATPase of plasmid R1, called ParM in this system, acts as a cytoskeletal element. The structure of ParM resembles eukaryotic actin and bacterial MreB protein and polymerizes into filamentous structures in the presence of ATP. In the R1 system, the partition site is called parC and the ParB-like protein is called ParR. Binding of ParM to the ParR/parC partition complexes stimulates the polymerization of ParM between complexes on daughter plasmids, effectively pushing the plasmids apart and to opposite ends of the dividing cell (see FIGURE 2.).

FIGURE 2. The partition of plasmid R1 involves polymerization of the ParM ATPase between plasmids.
In the other, nonactin class of partition ATPases, it is not known how these ParA proteins work to position plasmids. There are no sequences or structural similarities with ParM. It is possible that ParA proteins of plasmids such as P1 and F also act via polymerization. These ParA proteins do share some sequence similarities with the MinD ATPase that helps position the septum .
Intriguingly, some ParAs have been shown to oscillate over the bacterial nucleoid. The role of this oscillation is still a mystery, but these properties suggest that dynamic behavior of the ParA proteins is necessary for the partition reaction.
Proteins related to ParA and ParB are found in several bacteria. In Bacillus subtilis, they are called Soj and Spo0J, respectively. Mutations in these loci prevent sporulation because of a failure to segregate one daughter chromosome into the forespore. Mutations in the spo0J gene cause a 100-fold increase in the frequency of anucleate cells in vegetatively growing cells, suggesting that wildtype Spo0J contributes to chromosome segregation in normal cell cycles as well as during sporulation. Spo0J binds to a parS sequence that is present in multiple copies that are dispersed over about 20% of the chromosome in the vicinity of the origin. It is possible that Spo0J binds both old and newly synthesized origins, maintaining a status equivalent to chromosome pairing until the chromosomes are segregated to the opposite poles. In Caulobacter crescentus, ParA and ParB localize to the poles of the bacterium and ParB binds sequences close to the origin, thus localizing the origin to the pole. These results suggest that a specific apparatus is responsible for localizing the origin to the pole. The next stage of the analysis will be to identify the cellular components with which this apparatus interacts.
The importance to the plasmid of ensuring that all daughter cells gain replica plasmids is emphasized by the existence of multiple, independent systems in individual plasmids that ensure proper partition. Addiction systems, which operate on the basis of “we hang together or we hang separately,” ensure that a bacterium carrying a plasmid can survive only as long as it retains the plasmid.
There are several ways to ensure that a cell dies if it is “cured” of a plasmid, all of which share the principle illustrated in FIGURE 3. that the plasmid produces both a poison and an antidote. The poison is a killer substance that is relatively stable, whereas the antidote consists of a substance that blocks killer action but is relatively short lived. When the plasmid is lost the antidote decays, and then the killer substance causes the death of the cell. Thus, bacteria that lose the plasmid inevitably die, and the population is condemned to retain the plasmid indefinitely. These systems take various forms. One specified by the F plasmid consists of killer and blocking proteins. The plasmid R1 has a killer that is the mRNA for a toxic protein; the antidote is a small antisense RNA that prevents expression of the mRNA.
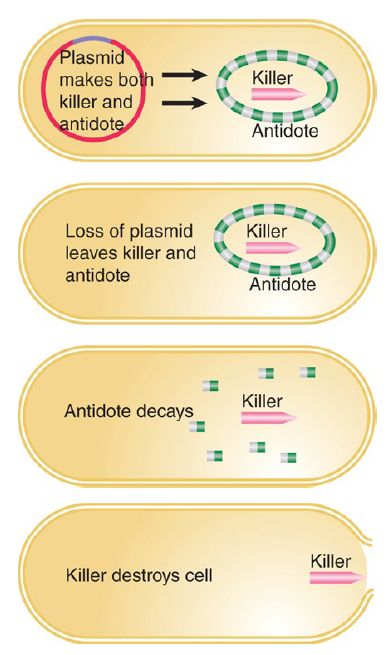
FIGURE 3. Plasmids might ensure that bacteria cannot live without them by synthesizing a long-lived killer and a short-lived antidote.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















