

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Coordinating Synthesis of the Lagging and Leading Strands
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
5-4-2021
2765
Coordinating Synthesis of the Lagging and Leading Strands
KEY CONCEPTS
- Different enzyme units are required to synthesize the leading and lagging strands.
- In E. coli, both of these units contain the same catalytic subunit (DnaE).
- In other organisms, different catalytic subunits might be required for each strand.
Each new DNA strand, leading and lagging, is synthesized by an individual catalytic unit. FIGURE 1. shows that the behavior of these two units is different because the new DNA strands are growing in opposite directions. One enzyme unit is moving in the same direction as the unwinding point of the replication fork and synthesizing the leading strand continuously. The other unit is moving “backward” relative to the DNA, along the exposed single strand. Only short segments of template are exposed at any one time. When synthesis of one Okazaki fragment is completed, synthesis of the next Okazaki fragment is required to start at a new location approximately in the vicinity of the growing point for the leading strand. This requires that DNA polymerase III on the lagging strand disengage from the template, move to a new location, and be reconnected to the template at a primer to start a new Okazaki fragment.
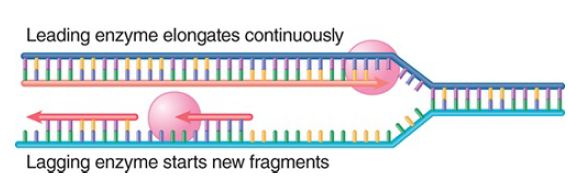
FIGURE 1. A replication complex contains separate catalytic units for synthesizing the leading and lagging strands.
The term enzyme unit avoids the issue of whether the DNA polymerase that synthesizes the leading strand is the same type of enzyme as the DNA polymerase that synthesizes the lagging strand. In the case we know best, E. coli, there is only a single DNA polymerase catalytic subunit used in replication, the DnaE polypeptide. Some bacteria and eukaryotes have multiple replication DNA polymerases .
The active replicase is an asymmetrical dimer with one unit on the lagging strand and one on the leading strand . Each half of the dimer contains DnaE as the catalytic subunit. DnaE is supported by other proteins (which differ between the leading and lagging strands).
The use of a single type of catalytic subunit, however, might be atypical. In the bacterium Bacillus subtilis, there are two different catalytic subunits. PolC is the homolog to E. coli’s DnaEBS and is responsible for synthesizing the leading strand. A related protein, DnaE is the catalytic subunit that synthesizes the lagging strand.
Eukaryotic DNA polymerases have the same general structure, with different enzyme units synthesizing the leading and lagging strands .
A major problem of the semidiscontinuous mode of replication follows from the use of different enzyme units to synthesize each new DNA strand: How is synthesis of the lagging strand coordinated with synthesis of the leading strand? As the replisome moves along DNA, unwinding the parental strands, one enzyme unit elongates the leading strand. Periodically, the primosome activity initiates an Okazaki fragment on the lagging strand, and the other enzyme unit must then move in the reverse direction to synthesize DNA. The next sections describe how leading and lagging strand replication is coordinated by interactions between the leading and lagging strand enzyme units.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















