

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
DNA Is Organized in Arrays of Nucleosomes
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
24-3-2021
2315
DNA Is Organized in Arrays of Nucleosomes
KEY CONCEPTS
-MNase cleaves linker DNA and releases individual nucleosomes from chromatin.
-More than 95% of the DNA is recovered in nucleosomes or multimers when MNase cleaves DNA in chromatin.
-The length of DNA per nucleosome varies for individual tissues or species in a range from 154 to 260 bp.
-Nucleosomal DNA is divided into the core DNA and linker DNA depending on its susceptibility to MNase.
-The core DNA is the length of 145–147 bp that is found on the core particles produced by prolonged digestion with MNase.
-Linker DNA is the region of 7 to 115 bp that is susceptible to early cleavage by nucleases.
When interphase nuclei are suspended in a solution of low ionic strength, they swell and rupture to release fibers of chromatin.
FIGURE .1 shows a lysed nucleus in which fibers are streaming out. In some regions, the fibers consist of tightly packed material, but in regions that have become stretched, they consist of discrete particles. These are the nucleosomes. In especially extended regions, individual nucleosomes are visibly connected by a fine thread, which is a free duplex of DNA. A continuous duplex thread of DNA runs through the series of particles.
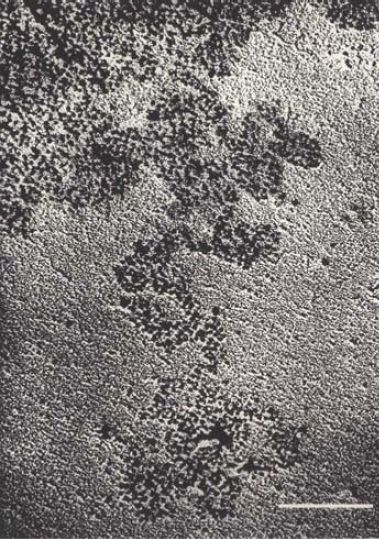
FIGURE.1 Chromatin spilling out of lysed nuclei consists of a compactly organized series of particles. The bar is 100 nm.
Reprinted from: Oudet, P., et al. 1975. “Electron microscopic and biochemical evidence.” Cell, 4:281–300, with permission from Elsevier (http://www.sciencedirect.com/science/journal/00928674). Photo courtesy of Pierre Chambon, College of France.
Researchers can obtain individual nucleosomes by treating chromatin with the endonuclease micrococcal nuclease (MNase), which cuts the DNA between nucleosomes, a region known as linker DNA. Ongoing digestion with MNase releases groups of particles, and eventually single nucleosomes. FIGURE .2 shows individual nucleosomes as compact particles measuring about 10 nm in diameter.

FIGURE .2 Individual nucleosomes are released by digestion of chromatin with micrococcal nuclease. The bar is 100 nm.
Reprinted from: Oudet, P., et al. 1975. “Electron microscopic and biochemical evidence.” Cell, 4:281–300, with permission from Elsevier (http://www.sciencedirect.com/science/journal/00928674). Photo courtesy of Pierre Chambon, College of France.
When chromatin is digested with MNase, the DNA is cleaved into integral multiples of a unit length. Fractionation by gel electrophoresis reveals the “ladder” presented in FIGURE .3.
Such ladders extend for multiple steps (about 10 are distinguishable in this figure), and the unit length, determined by the increments between successive steps, averages about 200 bp.
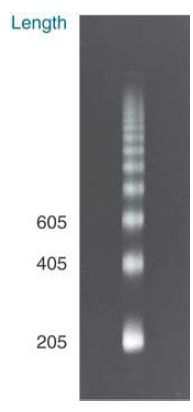
FIGURE .3 Micrococcal nuclease digests chromatin in nuclei into a multimeric series of DNA bands that can be separated by gel electrophoresis.
Photo courtesy of Markus Noll, Universität Zürich.
FIGURE .4 shows that the ladder is generated by groups of nucleosomes. When nucleosomes are fractionated on a sucrose gradient, they give a series of discrete peaks that correspond to monomers, dimers, trimers, and so on. When the DNA is extracted from the individual fractions and electrophoresed, each fraction yields a band of DNA whose size corresponds with a step on the micrococcal nuclease ladder. The monomeric nucleosome contains DNA of the unit length, the nucleosome dimer contains DNA of twice the unit length, and so on. More than 95% of nuclear DNA can be recovered in the form of the 200-bp ladder, indicating that almost all DNA must be organized in nucleosomes.
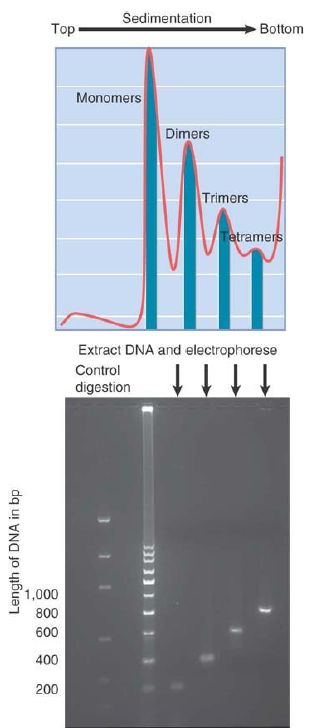
FIGURE .4 Each multimer of nucleosomes contains the appropriate number of unit lengths of DNA. In the photo, artificial bands simulate a DNA ladder that would be produced by MNase digestion. The image was constructed using PCR fragments with sizes corresponding to actual band sizes.
Photo courtesy of Jan Kieleczawa, Wyzer Biosciences.
The length of DNA present in the nucleosome can vary from the “typical” value of 200 bp. The chromatin of any particular cell type has a characteristic average value (±5 bp). The average most often is between 180 and 200, but there are extremes as low as 154 bp (in a fungus) or as high as 260 bp (in sea urchin sperm). The average value might be different in individual tissues of the adult organism, and there can be differences between different parts of the genome in a single cell type. Variations from the genome average often include tandemly repeated sequences, such as clusters of 5S RNA genes.
A common structure underlies the varying amount of DNA that is contained in nucleosomes of different sources. The association of DNA with the histone octamer forms a core particle containing 145–147 bp of DNA, irrespective of the total length of DNA in the nucleosome. The variation in total length of DNA per nucleosome is superimposed on this basic core structure.
The core particle is defined by the effects of MNase on the nucleosome monomer. The initial reaction of the enzyme is to cut the easily accessible DNA between nucleosomes, but if it is allowed to continue after monomers have been generated, it proceeds to digest some of the DNA of the individual nucleosome, as shown in FIGURE .5. Initial cleavage results in nucleosome monomers with (in this example) about 200 bp of DNA. After the first step, some monomers are found in which the length of DNA has been “trimmed” to about 165 bp. Finally, this is reduced to the length of the DNA of the core particle, 145–147 bp. After this, the core particle is resistant to further digestion by MNase.
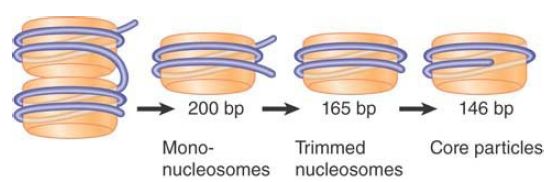
FIGURE .5 Micrococcal nuclease initially cleaves between nucleosomes. Mononucleosomes typically have ~200 bp DNA. Endtrimming reduces the length of DNA first to ~165 bp, and then generates core particles with 145–147 bp.
As a result of this type of analysis, nucleosomal DNA is functionally divided into two regions:
- Core DNA has a length of 145–147 bp, the length of DNA needed to form a stable monomeric nucleosome, and is relatively resistant to digestion by nucleases.
- Linker DNA comprises the rest of the repeating unit. Its length varies from as little as 7 bp to as many as 115 bp per nucleosome.
Core particles have properties similar to those of the nucleosomes themselves, although they are smaller. Their shape and size are similar to those of nucleosomes; this suggests that the essential geometry of the particle is established by the interactions between DNA and the protein octamer in the core particle. Core particles are readily obtained as a homogeneous population, and as a result they are often used for structural studies in preference to nucleosome preparations.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















