
Prokaryotic Gene Numbers Range Over an Order of Magnitude
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 11-3-2021
11-3-2021
 2052
2052
Prokaryotic Gene Numbers Range Over an Order of Magnitude
KEY CONCEPT
-The minimum number of genes for a parasitic prokaryote is about 500; for a free-living nonparasitic prokaryote, it is about 1,500.
Large-scale efforts have now led to the sequencing of many genomes. The range of known genome sizes (as summarized in TABLE 5.1) extends from the 0.6 × 106 base pairs (bp) of a mycoplasma to the 3.3 × 109 bp of the human genome, and includes several important model organisms, such as yeasts, the fruit fly, and a nematode worm. Many plant genomes are much
larger; the genome of bread wheat (Triticum aestivum L.) is 17 gigabases (Gb; five times the size of the human genome), though it should be noted that the species is hexaploid.
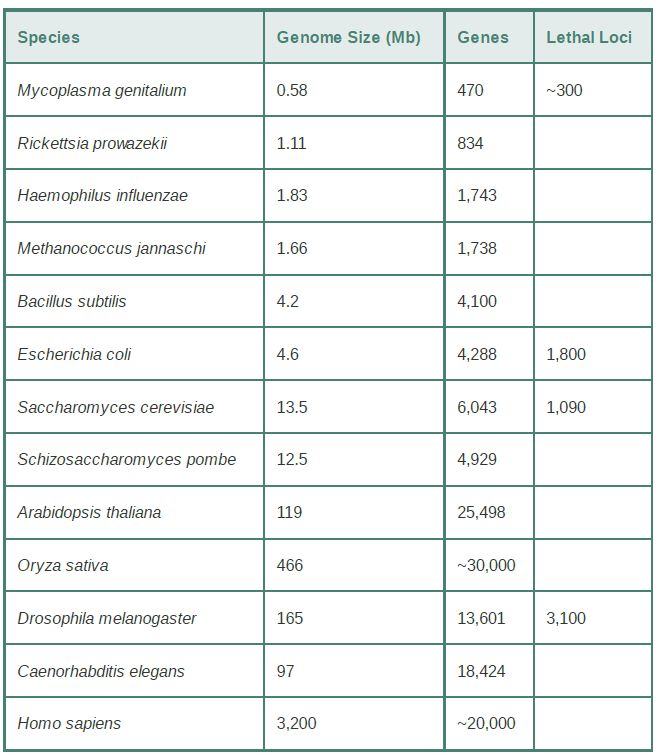
TABLE .1 Genome sizes and gene numbers are known from complete sequences for several organisms. Lethal loci are estimated from genetic data.
The sequences of the genomes of prokaryotes show that most of the DNA (typically 85% to 90%) encodes RNA or polypeptide.
FIGURE 1 shows that the range of prokaryotic genome sizes is an order of magnitude and that the genome size is proportional to the number of genes. The typical gene averages just under 1,000 bp in length.
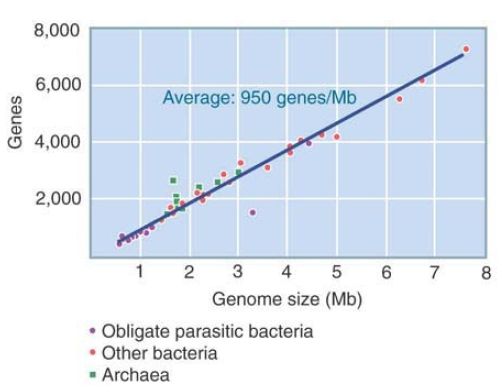
FIGURE 1 The number of genes in bacterial and archaeal genomes is proportional to genome size.
All of the prokaryotes with genome sizes below 1.5 Mb are parasites—they can live within a eukaryotic host that provides them with small molecules. Their genome sizes suggest the minimum number of functions required for a cellular organism. All classes of genes are reduced in number compared to prokaryotes with larger genomes, but the most significant reduction is in loci that encode enzymes involved with metabolic functions (which are largely provided by the host cell) and with regulation of gene expression. Mycoplasma genitalium has the smallest genome, with about 470 genes.
Archaeans have biological properties that are intermediate between those of other prokaryotes and those of eukaryotes, but their genome sizes and gene numbers fall in the same range as those of bacteria. Their genome sizes vary from 1.5 to 3 Mb, corresponding to 1,500 to 2,700 genes. Methanococcus jannaschii is a methane-producing species that lives under high pressure and temperature. Its total gene number is similar to that of Haemophilus influenzae, but fewer of its genes can be identified on the basis of comparison with genes known in other organisms.
Its apparatus for gene expression resembles that of eukaryotes more than that of prokaryotes, but its apparatus for cell division better resembles that of prokaryotes.
The genomes of archaea and the smallest free-living bacteria suggest the minimum number of genes required to make a cell able to function independently in its environment. The smallest archaeal genome has approximately 1,500 genes. The free-living nonparasitic bacterium with the smallest known genome is the thermophile Aquifex aeolicus, with a 1.5-Mb genome and 1,512 genes. A “typical” Gram-negative bacterium, H. influenzae, has 1,743 genes, the average size of which is about 900 bp. So, we can conclude that about 1,500 genes are required by an exclusively free-living organism.
Prokaryotic genome sizes extend over about an order of magnitude, from 0.6 Mb to less than 8 Mb. As expected, the larger genomes have more genes. The prokaryotes with the largest genomes, Sinorhizobium meliloti and Mesorhizobium loti, are nitrogen-fixing bacteria that live on plant roots. Their genome sizes (about 7 Mb) and total gene numbers (more than 7,500) are similar to those of yeasts.
The size of the genome of E. coli is in the middle of the range for prokaryotes. The common laboratory strain has 4,288 genes, with an average length of about 950 bp and an average separation between genes of 118 bp. There can be quite significan differences between strains, however. The known extremes in genome size among strains of E. coli are from 4.6 Mb with 4,249 genes to 5.5 Mb with 5,361 genes.
We still do not know the functions of all of these genes; functions have been identified for more than 80% of the genes. In most of these genomes, about 60% of the genes can be identified on the basis of homology with known genes in other species. These genes fall approximately equally into classes whose products function in metabolism, cell structure or transport of components, and gene expression and its regulation. In virtually every genome, 20% of the genes have not yet been ascribed any function. Many of these genes can be found in related organisms, which implies that they have a conserved function.
There has been some emphasis on sequencing the genomes of pathogenic bacteria, given their medical significance. An important insight into the nature of pathogenicity has been provided by the demonstration that pathogenicity islands are a characteristic feature of their genomes. These are large regions (from 10 to 200 kb) that are present in the genomes of pathogenic species but absent from the genomes of nonpathogenic variants of the same or related species. Their GC content often differs from that of the rest of the genome, and it is likely that these regions are spread among bacteria by a process of horizontal transfer. For example, the bacterium that causes anthrax (Bacillus anthracis) has two large plasmids (extrachromosomal DNA molecules), one of which has a pathogenicity island that includes the gene encoding the anthrax toxin.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


