

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Tools for Genetic Mapping
المؤلف:
John M Walker and Ralph Rapley
المصدر:
Molecular Biology and Biotechnology 5th Edition
الجزء والصفحة:
20-11-2020
2384
Tools for Genetic Mapping
1- Genetic Polymorphism
Some molecular variants are extremely common in populations; these commonly occurring variants are named polymorphisms and they have become one of the indispensable tools in genetics and genomics. Genetic polymorphism is a precise term which describes commonly occurring molecular variation at a specific locus such that at least 2% of the population will be heterozygous. ‘Locus’ means a DNA site in the genome, but not necessarily a functional gene. Most genetic polymorphisms have absolutely no effect on the phenotype of the organism, they represent ‘normal variation’.
2 - RFLPs and Minisatellites
The first DNA polymorphisms were detected using Southern blotting techniques of genomic DNA digested with restriction enzymes. This gave rise to restriction fragment length polymorphisms (RFLPs), which are due to the cleavage of certain alleles and the lack of cleavage of alternative alleles, resulting from the presence or absence of specific restriction enzyme recognition sites in the DNA. Southern blotting involves the transfer of DNA fragments generated by restriction enzyme digestion from an electrophoresis gel to an immobilising membrane such as nitrocellulose or nylon. The immobilised DNA is then probed with a radioactively or biotin-labelled DNA probe that could have originated from a cDNA clone or a genomic DNA clone or a PCR product. When the autoradiograph of the probed DNA digests from a set of unrelated individuals was examined, person to person variation in the resulting pattern of the bands identified an RFLP. The majority of RFLPs consist of two alleles (cut and uncut) and are found to result from single nucleotide replacements (Figure 1). Southern blotting remains one of the simplest methods to analyse gene dosage. The amount of the labelled probe that hybridises to the filter bound ‘target’ is proportional to the amount of target on the filter. Hence, when comparing the band patterns of samples that have been very carefully loaded on to the electrophoresis gel, it is possible to make a quantitative assessment using densitometry to measure the intensity of the bands on the X-ray film.
Restriction enzyme analysis of human DNA gave rise to a further class RFLPs known as VNTRs, where the variation is in the length of the DNA rather than the presence or absence of restriction enzyme sites. VNTRs are blocks of repetitive DNA sequences called minisatellites that occur throughout the genome, but that tend to be found towards the ends of human chromosomes. Hypervariable minisatellites were discovered by Jeffreys et al. in 1985 and were detected by hybridisation of a minisatellites probe to Southern blots of DNA digested with a restriction enzyme, usually HinF1, that cuts the DNA either side of the minisatellites. DNA probes such as 33.15 recognise a common core sequence of about 10–15 base pairs which was shared between many different minisatellite loci. These multilocus probes revealed tremendous person-to-person variations in the complex patterns of bands which became known as DNA fingerprints.36 DNA fingerprints were of great utility to forensic medicine and other fields where it was necessary to identify individuals unambiguously; however, DNA fingerprints were not amenable to mendelian segregation analysis or gene mapping because it was not possible to assign alleles within the complexity of bands displayed on the X-ray film.
A considerable advance with respect to gene mapping was made following the discovery of single-locus VNTR probes. Single-locus probes (SLP) were developed from specific cloned minisatellites and are thus able to detect individual minisatellite loci. They produce simple patterns on Southern blots in which heterozygous individuals display two bands and homozygotes a single band. Hence SLPs were useful for gene mapping since the bands could be tracked through families (Figure 2).

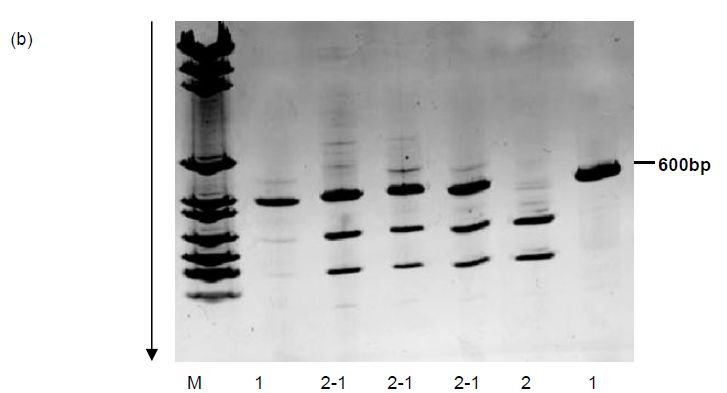
Figure 1. RFLP analysis of a 600 bp PCR product. (a) Schematic analysis of products from heterozygous and homozygous individuals digested with a restriction endonuclease. Three bands arise from the heterozygous individual, one which represents an uncleaved fragment and the other a cleaved fragment. (b) An SfaII RFLP revealed in six individuals following digestion of a 600 bp PCR product.
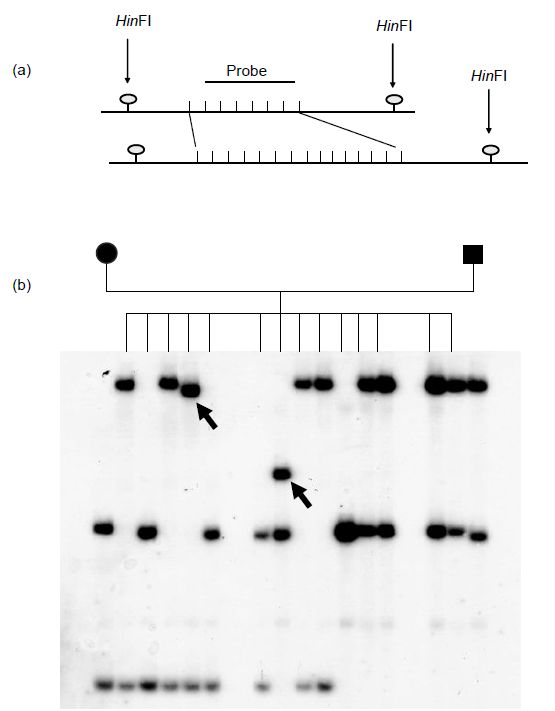
Figure 2. (a) Schematic illustration of a heterozygous VNTR locus. The invariant HinfI sites reveal a length polymorphism resulting from the repeat number variation in the two minisatellite alleles. (b) Southern blot analysis of hypervariable minisatellite B6.7 using a single locus probe on HinfI digests of genomic DNA from a large family. The arrows indicate new mutations not found in either parent.
The average heterozygosity of polymorphic VNTR loci is about 70%; those loci with very high heterozygosities, such as B6.7 in Figure 2, tend to exhibit considerable mutability and instability.37 It is estimated that there are between 15 000 and 20 000 VNTR loci in the human genome. However, as they tend to cluster preferentially near the ends of chromosomes, they are of limited use for genome mapping purposes.
Interestingly, it has since been discovered that the repeat blocks in many VNTR loci vary slightly within the sequences of the repeats, giving rise to repeat types along the length of the two alleles. These variants are detectable by PCR-based methods and they are the basis of the minisatellite variant repeats (MVR) system. This is a highly informative and elegant means of producing individual genetic barcodes which could be of tremendous use in forensic analysis of individuals in humans and many other species.
3- Microsatellites
Whereas minisatellites consisted of repeats of between 10 and 70, or more, DNA base pairs per repeat block, microsatellites are tandem repeats which have a much simpler structure, consisting of between two and five base pairs per repeat; they are also relatively stable. In contrast to minisatellites, microsatellites are amenable to PCR analysis because the overall length of the tandem repeat is relatively small, seldom exceeding more than a few hundred base pairs. Microsatellites have been widely used for gene mapping. In general, tri- and tetranucleotide repeats are preferred to dinucleotide repeat markers because they produce less confusing secondary bands (satellite bands) on electrophoresis gels. Microsatellites have been the most widely used marker polymorphisms in the Human Genome Project, where they have been used to build detailed linkage maps of all chromosomes; they also form the basis of modern forensic DNA profiling. Although the microsatellite blocks tend to be less polymorphic than the minisatellites in terms of heterozygosity, they are found scattered more evenly throughout the human genome rather than being concentrated in specific regions. The human genome is estimated to contain over one million such loci.Large-scale gene mapping studies frequently involve the analysis of numerous microsatellite loci by gel or capillary electrophoresis. This type of approach can yield several hundred genotypes from a single experiment. Capillary electrophoresis is amenable to automation such that the products of a PCR reaction can be introduced robotically to the electrophoretic instruments. In this case, the microsatellites are amplified using fluorophore-labelled PCR primers and the reaction products are detected by laser-stimulated fluorescence as they pass by a detector that records the electrophoresed DNA fragments. Automated techniques such as capillary electrophoresis can be carried out with relatively little technical support in comparison with the demands of gel electrophoresis. In practical terms, microsatellite analysis can be conduct edusing instruments such as the Applied Biosystems Genescan 3100. The dye-labelled PCR products are loaded on to the instrument and electrophoresed. Specialised software is then used to process and determine the allele sizes of the products and thus the genotypes. Interestingly, some triplet DNA repeat microsatellite sequences have been found to play a key role in the aetiology of some human genetic disorders, particularly those associated with certain inherited neurological conditions such as Huntington disease (HD, OMIM 143100), myotonic dystrophy (DM, OMIM 160900) and fragile X syndrome (FMR1, OMIM 309550). In these cases, the onset and severity of the disorders can be related to the increase in the number of trinucleotide repeats from generation to generation. For example, in HD, an autosomal dominant disease that gives rise to progressive and neural cell death, the onset of disease is associated with increases in the increasing length of a CAG triplet repeat located within the Huntington gene on human chromosome 4. The CAG triplet encodes the amino acid glutamine, which gives rise to a polyglutamine tract within the Huntington gene. It has been noted that in healthy individuals the number of CAGre peats ranges between 10 and 36, whereas in HD patients the number ranges between 37 and 100. The molecular mechanisms leading to microsatellite variation are not completely understood, although microsatellite loci certainly exhibit higher mutation rates compared with single nucleotide substitutions in genes. It is generally accepted that replication slippage is the most common mutational mechanism leading to the gain or loss of one or more repeat units. Other mutational mechanisms such as those based on unequal crossing over or duplication events have also been considered. The tendency to mutate might also depend on the chromosomal environment of a particular locus and whether or not it is transcribed.
4- Single Nucleotide Polymorphisms (SNPs)
By far the most widespread type of genetic variants is the single nucleotide polymorphism. Commonly referred to as SNPs (or Snips), these variant sites are found scattered throughout the genomes of most species. An SNP is caused by a single nucleotide replacement such as GAATTC to GTATTC that occurs with a frequency of at least 1% in the population. SNP is a new name for an old phenomenon. SNPs are the cause of most RFLPs and indeed the type of mutation that was indirectly detected in isozyme analysis by zone elctrophoresis before the advent of DNA technology.45 Detailed studies of human DNA suggest that SNPs are extremely abundant: a recent estimate is that there are 10 million SNPs in the human genome,46 which gives an average of one SNP per 300 base pairs. In a recent mapping effort, more than 3 million human SNPs have been analysed in more that 200 individuals. However, SNPs are not distributed evenly throughout the genome and the frequency can vary up to 10-fold.48 Unsurprisingly, because of selection pressure, SNPs are less abundant in the coding regions than non-coding regions; other factors that affect their distribution are local recombination and mutation rates. The majority of SNPs are located in repeat regions which are difficult to analyse; even so, SNPs are the best choice of polymorphic markers that cover the whole genome. Thus it has been estimated that there are 5.3 million common SNPs, each with a frequency of 10–15%, accounting for the bulk of DNA differences. Such SNPs present themselves on average once every 600 base pairs within the human genome in non-coding DNA,50 hence the level of heterozygosity in the human genome resulting from SNPs is extraordinarily high. Many of the current the SNP detection methods can equally detect small insertions and deletions in the DNA. Although SNPs have a use for classical gene mapping by linkage analysis, they have attracted great interest as tools for large-scale association studies with the purpose of associating specific sequences and mutations with specific phenotypes. SNPs can be used simply as genetic markers to identify disease genes by linkage or association analysis, or alternatively the SNPs themselves may on rare occasions be the actual variants in DNA sequences that cause the phenotype. SNPs within genes, especially coding regions that lead to missense mutations, may well change the function, regulation or expression of a protein.52 SNPs have also been discovered in the splice sites of genes and these can result in variant protein products with different exon arrays. A number of SNPs have been discovered in the controlling region of genes and some of these are reported to affect expression and regulation of proteins. It should be emphasised thatmo st SNPs occur in non-coding sequences far away from functional genes are therefore unlikely to have any functional significance, unless by chance they were to affect the configuration of a significant motif such as a remote locus control region. SNP analysis is currently seen as the most likely approach to discovering genes and mutations that contribute to disease. The US National Center for Biotechnology Information (NCBI) curates and maintains an SNP database (dbSNP) which contains sequence details of
several million SNPs, together with short insertions and deletions, from a variety of species. The structural simplicity of SNPs compared with microsatellite renders them ideal for full-scale automation and unambiguous scoring of alleles. However, in contrast with the microsatellite loci, because most SNPs represent single base changes or small insertions or deletions, they can have only two alleles and thus their heterozygosity cannot exceed 0.5, whereas microsatellite in contrast may have a vast number of alleles at individual loci with heterozygosities sometimes approaching 1.0. It
should be noted that the low polymorphism information content ofSNP s compared with microsatellites is more than offset by the fact that SNPs are present in such great abundance throughout the genome.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















