
Gene Expression Is Associated with Demethylation
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 8-5-2021
8-5-2021
 1975
1975
Gene Expression Is Associated with Demethylation
KEY CONCEPT
- Demethylation at the 5′ end of the gene is necessary for transcription.
Methylation of DNA is one of several epigenetic regulatory events that influence the activity of a promoter. Methylation at the promoter usually prevents transcription, and those methyl groups must be removed in order to activate a promoter. This effect is well characterized at promoters for both RNA polymerase I and RNA polymerase II. In effect, methylation is a reversible regulatory event, though DNA methylation patterns can also be stably maintained over many cell divisions. DNA methylation can be triggered by modifications to histones that include deacetylation and protein methylation .
Methylation also occurs in a particular epigenetic phenomenon known as imprinting. In this case, modification occurs in sexspecific patterns in sperm or oocyte, with the result that maternal and paternal alleles are differentially expressed in the next generation (see the chapter titled Epigenetics II).
Methylation at promoters for RNA polymerase II occurs on the 5′ position of C (producing 5-methyl cytosine, or 5mC) at CG doublets (also referred to as CpG doublets) by two different classes of DNA methyltransferases. DNMT1 is a maintenance enzyme that methylates the new C in a methylated GC doublet after replication.
DNMT2 is an enzyme that initiates de novo methylation of an unmethylated GC doublet. Although DNA methylation has been understood for some time, the mechanism of demethylation has been mysterious. Recently, the role of TET (ten eleven translocation) enzymes in demethylation of mammalian DNA has been proposed. These enzymes were originally identified as being involved in epigenetic inheritance and can convert 5mC to 5-hydroxymethylcytosine as the first step in a DNA damage excision repair pathway. A somewhat different DNA repair mechanism is known to be used for demethylation in plants.
Classically, the distribution of methyl groups was examined by taking advantage of restriction enzymes that cleave target sites containing the CG doublet. Two types of restriction activity are
compared in FIGURE 1. These isoschizomers are enzymes that cleave the same target sequence in DNA, but have a different response to its state of methylation. It is now possible through direct DNA sequencing to determine the methylome, or pattern of 5mC at single-base resolution in an organism.
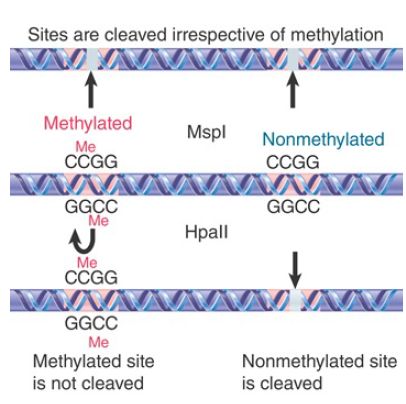
FIGURE 1. The restriction enzyme MspI cleaves all CCGG sequences whether or not they are methylated at the second C, but HpaII cleaves only unmethylated CCGG tetramers.
Many genes show a pattern in which the state of methylation is constant at most sites but varies at others. Some of the sites are methylated in all tissues examined; some sites are unmethylated in all tissues. A minority of sites are methylated in tissues in which the gene is not expressed, but are not methylated in tissues in which the gene is active. Even in active genes that are unmethylated in the promoter region these genes are typically methylated within the gene body, but usually not at the 3′ end. Thus, an active gene may be described as undermethylated.
Experiments with the drug 5-azacytidine produce indirect evidence that demethylation can result in gene expression. The drug is incorporated into DNA in place of deoxycytidine and cannot be methylated, because the 5′ position is blocked. This leads to the appearance of demethylated sites in DNA as the consequence of replication.
The phenotypic effects of 5-azacytidine include the induction of changes in the state of cellular differentiation. For example, muscle cells are induced to develop from non-muscle-cell precursors. The drug also activates genes on a silent X chromosome, which is consistent with the idea that the state of methylation is connected with chromosomal inactivity.
As well as examining the state of methylation of resident genes, we can compare the results of introducing methylated or nonmethylated DNA into new host cells. Such experiments show a clear correlation: The methylated gene is inactive, but the unmethylated gene is active.
What is the extent of the undermethylated region? In the chicken α- globin gene cluster in adult erythroid cells, the undermethylation is confined to sites that extend from about 500 bp upstream of the first of the two adult α genes to about 500 bp downstream of the second. Sites of undermethylation are present in the entire region, including the spacer between the genes. The region of undermethylation coincides with the region of maximum sensitivity to DNase I (see the Chromatin chapter). This argues that undermethylation is a feature of a domain that contains a transcribed gene or genes. As with many changes in chromatin, it seems likely that the absence of methyl groups is associated with the ability to be transcribed rather than with the act of transcription itself.
The problem in interpreting the general association between undermethylation and gene activation is that only a minority (sometimes a small minority) of the methylated sites are involved. It is likely that the state of methylation is critical at specific sites or in a restricted region. It is also possible that a reduction in the level of methylation (or even the complete removal of methyl groups from some stretch of DNA) is part of some structural change needed to permit transcription to proceed.
In particular, demethylation at the promoter may be necessary to make it available for the initiation of transcription. In the γ-globin gene, for example, the presence of methyl groups in the region around the start point, between −200 and +90, suppresses transcription. Removal of the three methyl groups located upstream of the start point, or of the three methyl groups located downstream, does not relieve the suppression. Removal of all methyl groups, though, allows the promoter to function. Transcription may therefore require a methyl-free region at the
promoter ( CpG Islands Are Regulatory Targets). There are exceptions to this general relationship. Some genes, however, can be expressed even when they are extensively methylated. Any connection between methylation and expression thus is not universal in an organism, but the general rule is that methylation prevents gene expression, and demethylation is required for expression.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


